Existence of virulence genes in clinical Shigella sonnei isolates from Jiangsu Province of China: a multicenter study
Introduction
Shigellosis is an acute invasive enteric infection caused by any of the four species of Shigella (S. dysenteriae, S. flexneri, S. boydii, and S. sonnei). S. flexneri is the most commonly isolated species in many developing countries (1,2), but S. sonnei in developed countries (3,4). With the development in China, S. sonnei plays an increasingly important part in Shigellosis (5). What’s more, the control of S. sonnei is inseparable from the research of the bacteria, including resistance, epidemiology, and virulence gene characteristics.
Although these were many studies involved the prevalence and antimicrobial resistance of S. sonnei from different parts of the world and China, little report investigated virulence genes of S. sonnei in the worldwide. Virulence factors, however, contribute to colonization and invasion of epithelial cells and eventually death of host cells. Different distribution of virulence genes in Shigella might cause different clinical manifestations (6,7). Invasion plasmid antigen H (ipaH) and invasion associated locus (ial) are responsible for the invasion of Shigella spp (8). Virulence genes encoded Shigella enterotoxin including Shigella enterotoxin 1 (ShET-1) and Shigella enterotoxin 2 (ShET-2). virF and virB (InvE) are two plasmid-borne proteins that control the expression of invasion genes (9). Finally, serin protease autotransporters of enterobacteriaceae (SPATEs), which has two phylogenetical classes, are present in Shigella spp. Secreted autotransporter toxin (sat) and Shigella IgA-like protease homologue (sigA) as two members of Class 1 are toxic to epithelial cells. pic (mucinase involved in colonization) and sepA as two members of Class 2 are non-toxic (10). The present study objects to investigate the prevalence and distribution of 11 virulence genes on S. sonnei isolated from patients with diarrhea in Jiangsu for the purpose of an epidemiological study.
Methods
A CDC-based active surveillance program was conducted in 13 cities of Jiangsu province from 2010 to 2015. Dysentery or diarrhea patients suspected of Shigella spp infection attending in different hospitals were enrolled for this study. Isolated samples were examined for Shigella spp. at each hospital using routine biochemical techniques. All collected isolates were further confirmed by Rapid ID32E strips (bioMérieux Corp., Singapore) and an automatic biochemistry analyzer (Hitachi 917; Boehringer Mannheim, Japan). By used of slide agglutination with hyperimmune sera (Ningbo Tianrong Bio-pharmaceutical Company Limited), O and H antigens were identified.
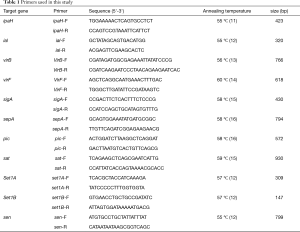
DNA extraction was performed using Qiagen DNA mini kit according to the manufacturer’s protocol. PCR was performed to target virulence genes by using previously reported primers (Table 1). Green Taq Mix (Vazyme, Nanjing, China) reaction was carried out according to the manufacturer’s instructions. The species were amplified under the following cycling conditions: initial denaturation at 95 °C for 5 min, followed by 30 cycles including denaturation for 50 s at 95 °C, annealing for 45 s (annealing temperature is shown in Table 1) and 72 °C for 1 min and a single final extension at 72 °C for 7 min. A representative amplicon was sequenced for each gene to validate that the primers amplified the target genes.
Full table
Statistical analyses were performed by using the database software program SPSS 16.0. Distribution of different virulence genes in serotypes, periods and regions were analyzed by Chi-square test. Statistical significance was set at P<0.05.
Results
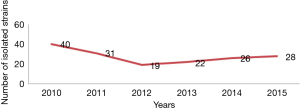
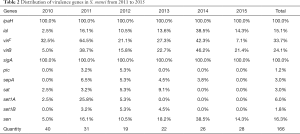
In the 6 years of the collection, A total of 166 strains of Shigella were collected (Figure 1). The prevalence of virulence genes among S. sonnei was shown in Table 2. All isolates were positive for ipaH gene, while justly 25 (15.1%) of the isolates were positive for ial gene in the present study (Table 2). a total of 40 (24.1%) and 56 (33.7%) isolates were found to be positive for virB and virF genes, respectively, and 30 (18.1%) strains found both virF and virB. All S. sonnei isolates harbored at least one SPATE proteins. The most common SPATE among S. sonnei strains was sigA (100% of strains), but another Class I SPATE, sat, was just existence in 5 strains of S. sonnei. The two Class 2 SPATEs, sepA and pic, were existence in 5 and 2 strains of S. sonnei respectively. The set1A gene was present in 10 (6.0%) S. sonnei isolates, and set1B was present in 3 (1.8%) S. sonnei isolates. Both set1A and set1B were detected in 2 (1.2%) strains of S. sonnei. The sen was present in 27 (16.3%) S. sonnei isolates. Interestingly, just one stain was positive for all virulence genes. In addition, the existence of virulence genes in S. sonnei changed in years (Table 2).
Full table
Discussion
Shigella remains to be the hallmark etiology of inflammatory diarrhea and dysentery and presents a serious challenge to public health, especially in developing countries and regions with substandard hygiene and poor quality water supplies. During the 6 years of this study, there was outbreak of S. sonnei in Jiangsu every year, and the numbers of isolated S. sonnei increased year by year after 2012, which showed a challenge for controlling infection of Shigella.
Multiple copies on large plasmid and chromosome may explain the ipaH gene being tested positive in all strains. Studies detected Shigella by a PCR assay targeting the ipaH gene, which found that the positive rate is higher than traditional culture method (17,18), and the present research confirmed ipaH is an appealing target for a diagnostic tool for it remains detectable even in the absence of the plasmid. Unlike ipaH gene, the ial gene located only on inv plasmid which was easily lost. The positive rate of ial gene in S. sonnei of Jiangsu was slightly lower than that in other regions (19,20). It should be noted that the existence of ial gene in S. sonnei was significantly lower than that in S. flexneri (2,21,22). The ial gene was involved in the invasion of intestinal cells (23), and the lower positive rate of this gene in S. sonnei might indicate lower aggressive.
When the growing condition is favorable for invasion, a transcriptional cascade is then initiated by activating virF gene to express the AraC-like protein virF, which in turn activates the transcription of the virB regulatory gene. The gene product virB protein consequently relieves the heat-stable nucleoid structural protein (H-NS) mediated transcriptional repression and activates the virulence genes on the plasmid-transcription of the virulence genes of Shigella is downregulates by H-NS in unfavorable growing condition (9,24). However, there were only 30 (18.1%) strains found both virF and virB. The low positive rate of those genes indicated that this classic pathway of regulating the expression of Shigella virulence gene does not play a major role in S. sonnei, and there might be other pathways for regulating gene expression.
There is species specificity in the distribution of SPATE. The high presence of sigA gene indicated sigA toxin may play an important role in the pathogenesis of S. sonnei strains, which was agreed with the previous article (20,22). For another class I SPATE, sat, the positive rate of the gene in S. sonnei was significantly lower than that of the gene in S. flexneri (6,25). Probably sat toxin has a major contribution in the virulence of S. flexneri strains. Similar to sat, the class II SPATEs (pic and sepA) might haven’t a significant effect on the pathogenicity of S. sonnei.
Shigella enterotoxin 1 (ShET-1) and ShET-2 could alter electrolyte and water transport in the small intestine, which could cause diarrhea and dehydration. ShET-1 is encoded in the set1 (A and B subunit) chromosomal gene that were almost exclusively found in S. flexneri isolates and rarely in other serotypes (26). Plasmid-encoded ShET-2 (encoded by sen) has been reported in different species of Shigella (27). the distribution of Shigella enterotoxin in S. sonnei was significantly lower than that in S. flexneri (21,26), which might mean that there is less danger of S. sonnei than S. flexneri.
In conclusion, this study provides baseline information on the distribution of virulence genes in clinical S. sonnei trains in Jiangsu province in China. Low distributions of genes encoding virulence factors in S. sonnei clinical isolates have been found compared with S. flexneri. The results obtained in this work contributed to a comprehensive understanding of the epidemiological status and characteristic of S. sonnei strains in Jiangsu Province.
Acknowledgments
We are very grateful to the provincial CDC and city CDC staff for their efforts to collect strains.
Funding: This work was supported by the National Natural Science Foundation of China (81471994), Jiangsu Provincial Natural Science Foundation (BK20170252), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six talent peaks project of Jiangsu Province (WSN-135), Advanced health talent of six-one project of Jiangsu Province (LGY2016042).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Alemu A, Geta M, Taye S, et al. Prevalence, associated risk factors and antimicrobial susceptibility patterns of infections among diarrheic pediatric population attending at Gondar town healthcare institutions, Northwest Ethiopia. Trop Dis Travel Med Vaccines 2019;5:7. [Crossref] [PubMed]
- Lanata CF, Fischer-Walker CL, Olascoaga AC, et al. Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLoS One 2013;8:e72788. [Crossref] [PubMed]
- Graciaa DS, Cope JR, Roberts VA, et al. Outbreaks Associated with Untreated Recreational Water - United States, 2000-2014. MMWR Morb Mortal Wkly Rep 2018;67:701-6. [Crossref] [PubMed]
- Lima IF, Havt A. Update on molecular epidemiology of Shigella infection. Curr Opin Gastroenterol 2015;31:30-7. [Crossref] [PubMed]
- Qian H, Liu G, Chen Y, et al. Increasing clinical resistance rate of to cefotaxime in Jiangsu Province, China, between 2012 and 2015. Ann Transl Med 2018;6:207. [Crossref] [PubMed]
- Niyogi SK, Vargas M. Prevalence of the sat, set and sen genes among diverse serotypes of Shigella flexneri strains isolated from patients with acute diarrhoea. Clin Microbiol Infect 2004;10:574-6. [Crossref] [PubMed]
- Sharma A, Singh SK. Phenotypic and genotypic characterization of Shigella spp. with reference to its virulence genes and antibiogram analysis from river Narmada. Indian J Microbiol 2009;49:259-65. [Crossref] [PubMed]
- Hazen TH, Leonard SR, Lampel KA, et al. Investigating the Relatedness of Enteroinvasive Escherichia coli to Other E. coli and Shigella Isolates by Using Comparative Genomics. Infect Immun 2016;84:2362-71. [Crossref] [PubMed]
- Jost BH. Site of transcriptional activation of virB on the large plasmid of Shigella flexneri 2a by VirF, a member of the AraC family of transcriptional activators. Microb Pathog 1993;14:481-8. [Crossref] [PubMed]
- Parham NJ, Pollard SJ, Desvaux M, et al. Distribution of the serine protease autotransporters of the Enterobacteriaceae among extraintestinal clinical isolates of Escherichia coli. J Clin Microbiol 2005;43:4076-82. [Crossref] [PubMed]
- Sangeetha AV, Parija SC, Mandal I. Clinical and microbiological profiles of shigellosis in children. J Health Popul Nutr 2014;32:580-6. [PubMed]
- Talukder KA, Mondol AS, Islam MA, et al. A novel serovar of Shigella dysenteriae from patients with diarrhoea in Bangladesh. J Med Microbiol 2007;56:654-8. [Crossref] [PubMed]
- Müller D, Greune L, Heusipp G, et al. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl Environ Microbiol 2007;73:3380-90. [Crossref] [PubMed]
- Vidal M, Kruger E, Durán C, et al. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J Clin Microbiol 2005;43:5362-5. [Crossref] [PubMed]
- Boisen N, Ruiz-Perez F, Scheutz F, et al. Short report: high prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am J Trop Med Hyg 2009;80:294-301. [Crossref] [PubMed]
- Restieri C, Garriss G, Locas MC. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl Environ Microbiol 2007;73:1553-62. [Crossref] [PubMed]
- Vu DT, Sethabutr O, Von Seidlein L, et al. Detection of Shigella by a PCR assay targeting the ipaH gene suggests increased prevalence of shigellosis in Nha Trang, Vietnam. J Clin Microbiol 2004;42:2031-5. [Crossref] [PubMed]
- Wang SM, Ma JC, Hao ZY, et al. Surveillance of shigellosis by real-time PCR suggests underestimation of shigellosis prevalence by culture-based methods in a population of rural China. J Infect 2010;61:471-5. [Crossref] [PubMed]
- Seribelli AA, Frazão MR, Medeiros MI. Molecular and phenotypic characterization of strains of Shigella sonnei isolated over 31 years suggests the circulation of two prevalent subtypes in São Paulo State, Brazil. J Med Microbiol 2016;65:666-77. [Crossref] [PubMed]
- Bakhshi B, Bayat B. Multiple- locus variable-number tandem-repeat analysis (MLVA) of Shigella sonnei isolates of 2012 outbreak I. R. Iran. Microb Pathog 2017;102:69-73. [Crossref] [PubMed]
- Hosseini Nave H, Mansouri S, Emaneini M. Distribution of genes encoding virulence factors and molecular analysis of Shigella spp. isolated from patients with diarrhea in Kerman, Iran. Microb Pathog 2016;92:68-71. [Crossref] [PubMed]
- da Cruz CB, de Souza MC, Serra PT, et al. Virulence factors associated with pediatric shigellosis in Brazilian Amazon. Biomed Res Int 2014;2014:539697. [PubMed]
- Vasconcellos FM, Tiba-Casas MR, Tavares LC, et al. Evaluation of a new trilocus sequence-based multiplex-PCR to detect major Acinetobacter baumannii clonal complexes circulating in Brazil. Infect Genet Evol 2017;54:4-6. [Crossref] [PubMed]
- Gao X, Zou T, Mu Z, et al. Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes. Nucleic Acids Res 2013;41:10529-41. [Crossref] [PubMed]
- Roy S, Thanasekaran K, Dutta Roy AR. Distribution of Shigella enterotoxin genes and secreted autotransporter toxin gene among diverse species and serotypes of shigella isolated from Andaman Islands, India. Trop Med Int Health 2006;11:1694-8. [Crossref] [PubMed]
- Vargas M, Gascon J, Jimenez De Anta MT. Prevalence of Shigella enterotoxins 1 and 2 among Shigella strains isolated from patients with traveler's diarrhea. J Clin Microbiol 1999;37:3608-11. [PubMed]
- Nataro JP, Seriwatana J, Fasano A, et al. Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infect Immun 1995;63:4721-8. [PubMed]