MiR-195 restrains lung adenocarcinoma by regulating CD4+ T cell activation via the CCDC88C/Wnt signaling pathway: a study based on the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and bioinformatic analysis
Introduction
Non-small cell lung cancer (NSCLC), which accounts for 85% of all lung cancer cases, is one of the leading causes of cancer-related deaths worldwide (1,2). More specifically, lung adenocarcinoma (LUAD) is the most common subtype of NSCLC. Despite improvements in early disease detection and the development of chemotherapeutic and targeted treatments, the overall survival rate of LUAD patients remains poor (2). In recent years, immunotherapy has attracted increasing attention from oncologists. T cells are important mediators of tumor immunity, and in most types of solid tumors, T cell infiltration is a favorable prognostic marker (3,4). Immunotherapy to boost T cell functionality in tumors is rapidly becoming established as a standard treatment (5), and the immunotherapy focus has been on recruiting tumor infiltrating T cells (6). CD4+ T cells secrete a variety of cytokines that have direct effector functions and activate other immune cells (such as B cells, dendritic cells and even CD8 T Cells) (7,8). In lung cancer, tumor-infiltrating CD4+ T cells plays an essential role in the immune response (9). CD4+ T cells affect tumors by allowing CD8+ T cells entry to tumor sites (10) and infected mucosa (11); furthermore, they are also required for the inhibition of angiogenesis at tumor sites (12).
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression by degrading or inhibiting translation of their target transcripts, thereby affecting processes such as cell proliferation, differentiation and apoptosis (13). Changes in miRNA expression were reported as biomarkers for LUAD risk and prognosis (14), and miRNA-based biomarkers with prognostic or predictive potential for tumor responsiveness to immunocheckpoint inhibitors were recently described (15). Cell-specific miRNA expression patterns and the roles of miRNAs in the LUAD microenvironment have not been fully elucidated. Patients with similar clinical features often have different outcomes, suggesting an underlying relationship between LUAD development and genetic variations. The identification of new specific biomarkers that can be used to monitor tumor progression and treatment sensitivity, as well as to predict patient survival, will help overcome these challenges and improve outcomes in LUAD patients (16,17).
Gene expression profiling has become a new and effective method to identify prognostic markers and molecular targets for therapies (18). Dysregulated miRNAs in LUAD can be identified using miRNA expression profiling. The aim of our study was to use bioinformatic analysis of a large clinical dataset to systematically identity microRNA signatures, as well as miRNA-gene axes, related to LUAD and to explore potential biomarkers and mechanisms associated with LUAD immune responses.
Methods
Microarray profiles from the Gene Expression Omnibus (GEO) database
LUAD-related microarray profiles (up to November 2018) were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The search criterion of GEO Databases was shown in Table S1. Microarrays that met the following criteria were collected: (I) studies including at least 60 samples and (II) examination of miRNA expression in both cancerous tissue and adjacent noncancerous tissue from LUAD patients. Microarrays without useful data for analysis were excluded. Differentially expressed miRNAs (DEMs) between LUAD cancerous tissue and adjacent noncancerous tissue samples in each GEO dataset were ranked by the Robust Multi-Array Average and Linear Models for Microarray package and annotated by converting the different probe IDs to gene IDs.

Full table
Integrated analysis of miRNA expression datasets
The RobustRankAggreg (RRA) package was used to identify DEMs between LUAD cancerous tissue and adjacent noncancerous tissue samples. The adjusted P value and Log2-fold change (FC) were specified as 0.05 and 1, respectively. One-sided test was applied to classify the downregulated DEMs. We selected the top 10 significantly downregulated DEMs for further studies.
miRNA-seq data from The Cancer Genome Atlas (TCGA) database
Publicly available miRNA-seq data on miRNA levels in LUAD cancer tissue and adjacent noncancerous tissue samples were directly downloaded from the TCGA data portal (http://cancergenome.nih.gov/). We obtained the miRNA profiles of 209 LUAD cancer tissue samples and 45 adjacent noncancerous tissue samples together with the clinical information (level 3) of the corresponding patients. DEMs between the LUAD samples with pathological stages of I–IV and adjacent noncancerous tissue samples were identified by calculating the FC (|log2(FC)| >2 and adjusted P value <0.05) with the R package edgeR.
Integrated analysis the GEO profiles and the TCGA miRNA-seq data
The top 10 DEMs identified as significantly downregulated in the GEO database were entered into the TCGA database for further verification. DEMs that showed consistent expression in GEO were selected for statistical analysis. Independent Student’s t-tests were performed to calculate the differences in the miRNA levels between LUAD cancerous tissue and adjacent noncancerous tissue. P<0.05 was considered statistically significant.
Diagnosis and prognosis analysis
A receiver operating characteristic (ROC) curve built on a univariate classification model based on the DEM expression profiles across independent TCGA datasets were used to predict LUAD. Kaplan-Meier plots of the overall survival for a discriminatory median DEM expression profile based on TCGA sequencing data were used to assess prognostic accuracy. P values were calculated using the log-rank test.
MiRNAs meeting the above diagnostic and prognostic criteria were introduced into multiple linear regression models for further analysis. The relative miRNA levels were treated as an independent variable, and the diagnosis results were treated as a dependent variable. A linear regression equation was constructed to identify miRNAs with independent diagnostic value.
Pairwise meta-analysis and diagnostic meta-analysis
A comprehensive meta-analysis was performed using Stata 14.0 software (Stata Corporation, College Station, TX), combining the TCGA data and GEO datasets. The pooled data in the meta-analysis were assessed by the standard mean difference (SMD) with a 95% confidential interval (CI). Heterogeneity among the eligible microarrays was evaluated by chi-squared and I-squared tests. The effect model was then determined according to the heterogeneity. Specifically, a fixed effects model was conducted for the meta-analysis when the heterogeneity was low (I2≤50% and P>0.1), and a random effects model was selected if apparent heterogeneity existed (I2>50% or P≤0.1). A bivariate-mixed model was used to estimate the ROC curve, and the area under curve (AUC) was also estimated to optimize cut-off points.
Target prediction and functional analysis of miRNA
The presumed targets of the integrated-signature miRNAs were identified by 3 different target prediction algorithms: TargetScan, miRDB and DIANA-TarBase. Unique genes with target sites in 3' UTR sequences were included. To assess the possible functions, we searched the Gene Ontology (GO) database, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and the Database for Annotation, Visualization and Integrated Discovery (DAVID). A P value less than 0.01 was defined as the cutoff criterion for KEGG pathways enriched in the target gene set.
Gene set enrichment analysis (GSEA)
The enrichment analyses for immunologic signature gene sets were conducted with GSEA v3.0 for the target genes. The enriched pathways were arranged in the order of their normalized enrichment scores (NESs).
Immunocyte infiltration in the tumor microenvironment
The core enriched genes have been packaged into the web-accessible resource TIMER (Tumor IMmune Estimation Resource; https://cistrome.shinyapps.io/timer/), to enable further exploration of the impacts of the core enriched genes on immunocyte infiltration in tumor microenvironments.
Results
Collection of microarray datasets from GEO
The flow chart for the study selection for this integrated analysis is shown in Figure 1. We searched the GEO database, and the GEO microarrays can be regarded as a training dataset to screen for DEMs in LUAD. Finally, 4 GEO datasets (accession numbers GSE48414, GSE51853, GSE63805 and GSE74190) were included in the present study, and the characteristics of the studies based on the GEO dataset are presented Table 1.
Full table
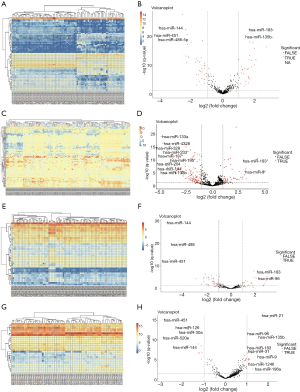
Due to the heterogeneity in the sample types in the GSE microarrays, the common DEMs were examined separately (Figure 2). The downregulated DEMs in each GSE are presented in http://fp.amegroups.cn/cms/atm.2019.05.54-1.pdf. Tables S2-S4. There were inconsistencies in the DEMs obtained from each GSE microarray. Therefore, the RRA package was used to perform an integrated analysis of the 4 GSE microarrays to identify co-downregulated DEMs. There were 27 significantly downregulated miRNAs. The hierarchical clustering of the top 10 miRNAs is shown in Figure 3.
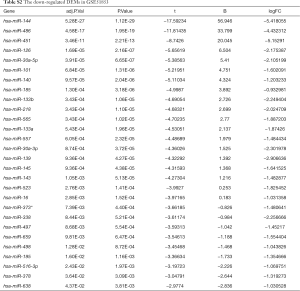
Full table
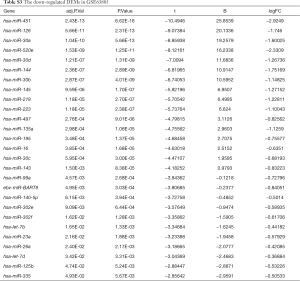
Full table
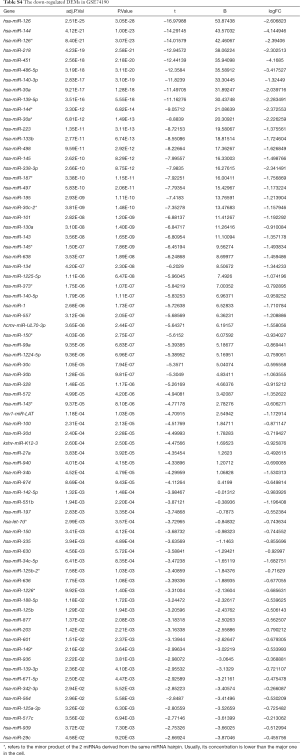
Full table
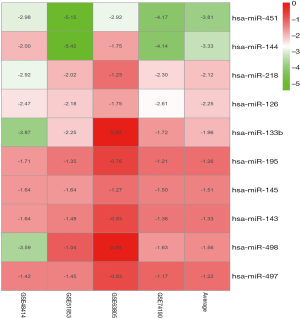
Integrated-signature miRNAs showed clinical prognostic significance in LUAD patients
We further validated the top 10 downregulated DEMs in TCGA-LUAD samples (209 LUAD cancerous tissue samples and 45 adjacent noncancerous tissue samples, http://fp.amegroups.cn/cms/atm.2019.05.54-2.pdf). Only miR-195, miR-451, miR-144, miR-218, miR-133b, miR-145, miR-143 and miR-497 were significantly downregulated in LUAD tumors (Figure 4). The diagnostic efficiency and prognostic value of each of these miRNAs were estimated via ROC curve analysis and Kaplan-Meier survival analysis, respectively. Ultimately, we selected 3 miRNAs (miR-143, miR-195 and miR-218) with high diagnostic efficiency (AUC >0.8, P<0.05) and prognostic value (logrank P<0.05) (Figure 5). We next optimized the accuracy by using a linear regression model built on a panel of the combined miRNAs. By constructing the linear regression equation LUAD risk score = −0.0.02267miR-143-0.1115miR-195-0.04098miR-218+2.3699, miR-195 was identified as the most significant independent variable (P=0.0006, Table 2).
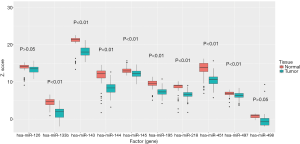
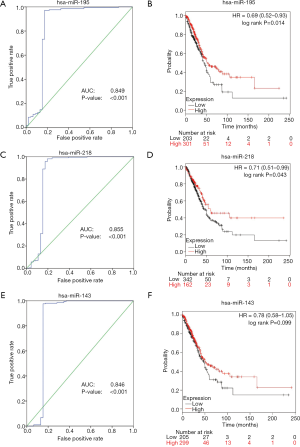

Full table
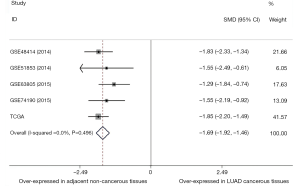
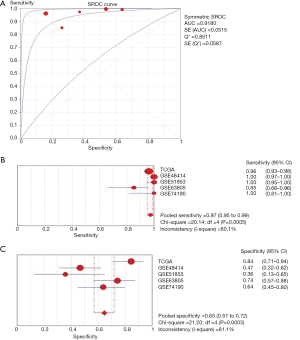
Combining the TCGA data and GEO datasets, the results of pairwise meta-analyses indicated that miR-195 was overexpressed in adjacent noncancerous tissue samples (SMD =−1.69, 95% CI: −1.92 to −1.46, Figure 6). Furthermore, the results of a diagnostic meta-analysis suggested that miR-195 offers high diagnostic efficiency (AUC =0.9180, Figure 7A; the pooled sensitivity =0.97, 95% CI: 0.95–0.99, Figure 7B; the pooled specificity =0.65, 95% CI: 0.57–0.72, Figure 7C).
Target gene prediction coupled with pathway analysis
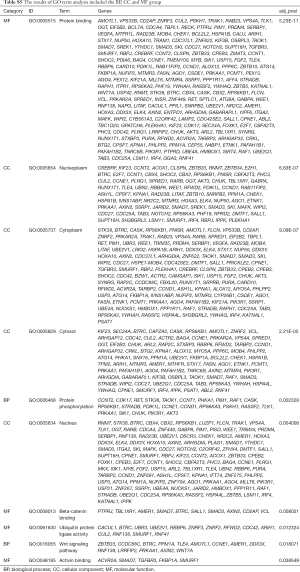
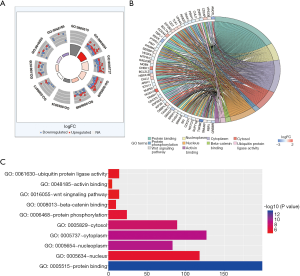
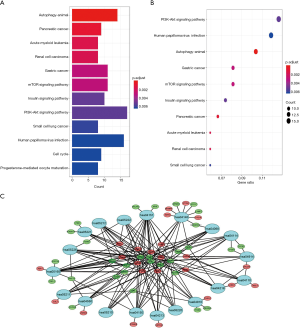
To explore the biological mechanisms of miR-195 in LUAD, we performed target gene prediction coupled with pathway analysis. A total of 287 target genes (http://fp.amegroups.cn/cms/atm.2019.05.54-3.pdf) were identified via TargetScan, miRDB and DIANA-TarBase, and these genes were then subjected to GO and KEGG analyses. The results of the GO term analysis included the biological process (BP), cellular component (CC) and molecular function (MF) groups. The target genes were mainly enriched in protein binding, beta-catenin binding, ubiquitin protein ligase activity and activin binding in the MF group; nucleoplasm, cytoplasm, cytosol and nucleus in the CC group; and protein phosphorylation and Wnt signaling pathway in the BP group (Table S5 and Figure 8). The results of the GO and KEGG analysis indicated that the most significantly enriched terms were “protein binding” and “cell cycle”. The top 50 genes with significant differences in their expression levels are shown along with their functions in Figure 8B. All of the target genes were analyzed using the KEGG pathway website and the clusterProfiler package of the R software, and only genes with P values less than 0.01 were included. The largest number genes were enriched in the PI3K-Akt signaling pathway (Figure 9). The specific links between each gene and its function are shown in Figure 9C.
Full table
GSEA in immunologic signature gene sets
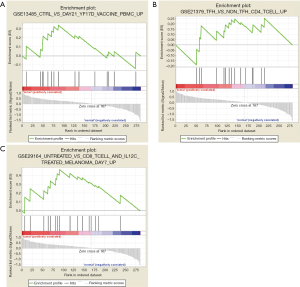
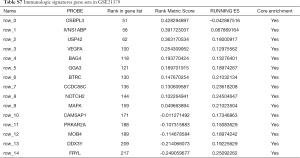
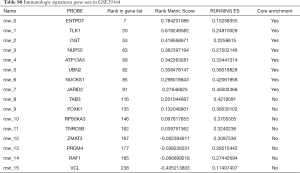
To characterize the potential mechanisms of immunologic function associated with the miR-195 target genes, GSEA was used to obtain the biological processes enriched in immunologic signature gene sets. Then, 3 functional gene sets were enriched (GSE13485, GSE21379 and GSE29164, Figure 10), and they were all upregulated in the tumor tissue samples. The core genes of the 3 immunologic signature gene sets are shown in Tables S6-S8.
Full table
Full table
Full table
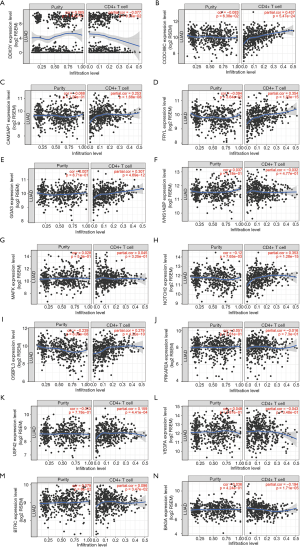
GSE21379 was associated with upregulation of CD4+ T cells in tumors, and the core enrichment genes were validated via the TIMER database. The correlations between the expression levels of 14 genes (OSBPL3, IVNS1ABP, USP42, VEGFA, BAG4, GGA3, BTRC, CCDC88C, NOTCH2, MAFK, CAMSAP1, PRKAR2A, MOB4, DDX3Y and FRYL) and CD4+ T cell infiltration were examined. The expression levels of CCDC88C were significantly correlated with CD4+ T cell activation (partial.cor =0.437, P<0.001, Figure S1).
Discussion
The present study, based on GEO and TCGA analysis, revealed that miR-195 was overexpressed in adjacent noncancerous tissue samples, and that it had high diagnostic efficiency. These results are consistent with those of a previous study (19) that showed that miR-195 suppressed tumor cell growth, migration and invasion and was associated with better survival outcomes in LUAD patients.
Nevertheless, most previous basic studies focused on one miRNA-195 target gene, i.e., CHEK1 (19), IRS1 (20), or MMP14 (21). Our KEGG pathway analysis found that the largest number genes were enriched in the PI3K-Akt signaling pathway, including the following genes: CCNE1, FGF2, PIK3R1, AKT3, RPS6KB1, PHLPP2, ITGA2, YWHAQ, YWHAH, CCND1, PRKAA1, MYB, RAF1, INSR, VEGFA, LAMC1 and CHUK. Because miRNAs are mainly negative regulators of their target genes, these upregulated genes (CCNE1, RPS6KB1, ITGA2, YWHAQ, PRKAA1, INSR, VEGFA, LAMC1 and CHUK) should be given attention in future studies of LUAD. The PI3K-Akt signaling pathway is essential for maintaining cell growth, survival, death and metabolism, and it is commonly activated during cancer initiation and progression (22). In addition, the PI3K-Akt signaling pathway can regulate the proliferation, migration, invasion, apoptosis and angiogenesis of lung cancer cells (23), and activation of the PI3K-Akt signaling pathway may be a therapeutic molecular target for lung cancer (24). In addition to the relationship between VEGFA and the PI3K-Akt signaling pathway in lung cancer (25), the biological behaviors of lung cancer involving the PI3K-Akt signaling pathway remain to be investigated.
The GSEA-based identification of an immunologic signature gene set was an important objective of this study. GSE13485 was mainly related to a vaccine response, and GSE29164 was based on data collected during immunotherapy for melanoma. Therefore, we focused on the relationship between the genes and immune processes contained in GSE21379. As shown in Table S7, 15 core genes (OSBPL3, IVNS1ABP, USP42, VEGFA, BAG4, GGA3, BTRC, CCDC88C, NOTCH2, MAFK, CAMSAP1, PRKAR2A, MOB4, DDX3Y and FRYL) were involved in the upregulation of CD4+ T cells in tumor tissue. Previous studies (26,27) showed that CD4+ T cells induced cytotoxic programming of CD8+ T cells, which then suppress tumor growth via IFN-γ secretion or direct killing of the tumor cells (28,29). However, the effects of CD4+ T cell infiltration on the biological behaviors of tumors are not consistent. The presence of CD4+ T cells in the tumor microenvironment was linked to poor outcomes in prostate cancer patients (30) as well as in patients with renal cell carcinoma (31). CD4+ T cells recruited in mammary cancer enhanced metastasis (32). Among the immunologic signature gene sets in GSE13485, NOTCH2 (33-35) and VEGFA (36) had significant effects on CD4+ T cells. However, the immune regulatory mechanisms of the other genes in tumors and lung cancer are not fully elucidated. Therefore, our study provides a clue for studying the genetic regulation of CD4+ T cells and lung cancer immunity.
Our results also demonstrated that the CCDC88C expression level was significantly correlated with CD4+ T cell activation. Enomoto et al. (37) found that CCDC88C (coiled-coil domain containing 88C) encodes a member of the hook-related proteins involved in the regulation of the Wnt signaling pathway. These results are consistent with our enrichment analysis. Furthermore, the Wnt signaling pathway controls inflammatory responses induced by multiple factors, such as pathogenic bacteria via Toll-like receptors (38,39), and it might be involved in the impaired T-cell homeostasis present in a variety of immune system diseases, such as rheumatoid arthritis and systemic lupus erythematosus (40). Inhibition of the Wnt signaling pathway enhanced CD4+ T cell infiltration into the central nervous system by increasing the expression of vascular cell adhesion molecule-1 and the transcytosis protein Caveolin-1, as well as by promoting endothelial transcytosis (41). A previous study (42) showed that both Wnt3a and β-catenin were overexpressed by tumor-infiltrating and nontumor-infiltrating CD4+ or CD8+ T cells. Wnt3a blockade inhibited the differentiation of naive T cells but could not rescue the dysfunction of differentiated T cells in the tumor environment. The canonical Wnt signaling pathway blocks T cell differentiation and plays an important role in phenotypic maintenance of naive T cells and stem cell-like memory T cells in human peripheral blood (43); however, its effects on tumor-infiltrating lymphocytes in non-small cell lung cancer are still unclear. Based on the results of our bioinformatic analysis and previous literature reports, we conclude that CCDC88C might regulate CD4+ T cell activation via the Wnt signaling pathway.
However, this conclusion should be treated with caution. The GO enrichment analysis showed that CCDC88 was enriched in the Wnt signaling pathway, but this pathway was not significantly enriched in the KEGG results. In general, the biological process results from the GO analysis have many similar functions to those identified via the KEGG pathway analysis. Since the two types of enrichment analysis are based on different databases, there may be some inconsistencies in the results. However, this inconsistency could represent a cross-complement that provides verification of the two methods.
The tumor microenvironment, with its individual immune cells, may play key roles in tumor progression. Cancer development is driven by the accumulation of random mutations that lead to increased dysregulation of several key pathways. Therefore, it is very important to use bioinformatics approaches to identify key genes that shape tumor immune microenvironments.
Acknowledgments
Funding: This work was supported by grants from the Chinese National Natural Science Foundation (grant No. 81572967, 81372498, and 81800429), Hubei Natural Science Foundation (grant No. 2013CFA006), and Zhongnan Hospital of Wuhan University Science, Technology and Innovation Seed Fund (grant No. znpy2016050, znpy2017001, and znpy2017049), National key clinical speciality construction program of China [No. (2013)544], Wuhan City Huanghe Talents Plan and the Fundamental Research Funds for the Central Universities (grant No. 2042018kf0065).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Blandin Knight S, Crosbie PA, Balata H, et al. Progress and prospects of early detection in lung cancer. Open Biol 2017;7:170070. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298-306. [Crossref] [PubMed]
- Becht E, Giraldo NA, Dieu-Nosjean MC, et al. Cancer immune contexture and immunotherapy. Curr Opin Immunol 2016;39:7-13. [Crossref] [PubMed]
- Oja AE, Piet B, van der Zwan D, et al. Functional Heterogeneity of CD4+ Tumor-Infiltrating Lymphocytes With a Resident Memory Phenotype in NSCLC. Front Immunol 2018;9:2654. [Crossref] [PubMed]
- Bruno TC, Ebner PJ, Moore BL, et al. Antigen-Presenting Intratumoral B Cells Affect CD4+ TIL Phenotypes in Non-Small Cell Lung Cancer Patients. Cancer Immunol Res 2017;5:898-907. [Crossref] [PubMed]
- Kamphorst AO, Ahmed R. CD4 T-cell immunotherapy for chronic viral infections and cancer. Immunotherapy 2013;5:975-87. [Crossref] [PubMed]
- Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4+ T cells in immunity to viruses. Nat Rev Immunol 2012;12:136-48. [Crossref] [PubMed]
- Hiraoka K, Miyamoto M, Cho Y, et al. Concurrent infiltration by CD8+ T cells and CD4+ T cells is a favourable prognostic factor in non-small-cell lung carcinoma. Br J Cancer 2006;94:275-80. [Crossref] [PubMed]
- Bos R, Sherman LA. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res 2010;70:8368-77. [Crossref] [PubMed]
- Nakanishi Y, Lu B, Gerard C, et al. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature 2009;462:510-3. [Crossref] [PubMed]
- Rakhra K, Bachireddy P, Zabuawala T, et al. CD4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell 2010;18:485-98. [Crossref] [PubMed]
- Pritchard CC, Kroh E, Wood B, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492-7. [Crossref] [PubMed]
- Świtlik WZ, Szemraj J. Circulating miRNAs as non-invasive biomarkers for non-small cell lung cancer diagnosis, prognosis and prediction of treatment response. Postepy Hig Med Dosw (Online) 2017;71:649-62. [Crossref] [PubMed]
- Halvorsen AR, Sandhu V, Sprauten M, et al. Circulating microRNAs associated with prolonged overall survival in lung cancer patients treated with nivolumab. Acta Oncol 2018;57:1225-31. [Crossref] [PubMed]
- Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer 2016;16:525-37. [Crossref] [PubMed]
- Neal JW, Gainor JF, Shaw AT. Developing biomarker-specific end points in lung cancer clinical trials. Nat Rev Clin Oncol 2015;12:135-46. [Crossref] [PubMed]
- Khan J, Wei JS, Ringnér M, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med 2001;7:673-9. [Crossref] [PubMed]
- Liu B, Qu J, Xu F, et al. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget 2015;6:9445-56. [PubMed]
- Wang Y, Zhang X, Zou C, et al. miR-195 inhibits tumor growth and angiogenesis through modulating IRS1 in breast cancer. Biomed Pharmacother 2016;80:95-101. [Crossref] [PubMed]
- Li M, Ren CX, Zhang JM, et al. The Effects of miR-195-5p/MMP14 on Proliferation and Invasion of Cervical Carcinoma Cells Through TNF Signaling Pathway Based on Bioinformatics Analysis of Microarray Profiling. Cell Physiol Biochem 2018;50:1398-413. [Crossref] [PubMed]
- Koundouros N, Poulogiannis G. Phosphoinositide 3-Kinase/Akt Signaling and Redox Metabolism in Cancer. Front Oncol 2018;8:160. [Crossref] [PubMed]
- Zhou Y, Li S, Li J, et al. Effect of microRNA-135a on Cell Proliferation, Migration, Invasion, Apoptosis and Tumor Angiogenesis Through the IGF-1/PI3K/Akt Signaling Pathway in Non-Small Cell Lung Cancer. Cell Physiol Biochem 2017;42:1431-46. [Crossref] [PubMed]
- Matsuda S, Nakagawa Y, Kitagishi Y, et al. Reactive Oxygen Species, Superoxide Dimutases, and PTEN-p53-AKT-MDM2 Signaling Loop Network in Mesenchymal Stem/Stromal Cells Regulation. Cells 2018;7:E36. [Crossref] [PubMed]
- Chen CH, Lai JM, Chou TY, et al. VEGFA upregulates FLJ10540 and modulates migration and invasion of lung cancer via PI3K/AKT pathway. PLoS One 2009;4:e5052. [Crossref] [PubMed]
- Ahrends T, Bąbała N, Xiao Y, et al. CD27 Agonism Plus PD-1 Blockade Recapitulates CD4+ T-cell Help in Therapeutic Anticancer Vaccination. Cancer Res 2016;76:2921-31. [Crossref] [PubMed]
- Ahrends T, Spanjaard A, Pilzecker B, et al. CD4+ T Cell Help Confers a Cytotoxic T Cell Effector Program Including Coinhibitory Receptor Downregulation and Increased Tissue Invasiveness. Immunity 2017;47:848-61.e5. [Crossref] [PubMed]
- Quezada SA, Simpson TR, Peggs KS, et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med 2010;207:637-50. [Crossref] [PubMed]
- Friedman KM, Prieto PA, Devillier LE, et al. Tumor-specific CD4+ melanoma tumor-infiltrating lymphocytes. J Immunother 2012;35:400-8. [Crossref] [PubMed]
- McArdle PA, Canna K, McMillan DC, et al. The relationship between T-lymphocyte subset infiltration and survival in patients with prostate cancer. Br J Cancer 2004;91:541-3. [Crossref] [PubMed]
- Bromwich EJ, McArdle PA, Canna K, et al. The relationship between T-lymphocyte infiltration, stage, tumour grade and survival in patients undergoing curative surgery for renal cell cancer. Br J Cancer 2003;89:1906-8. [Crossref] [PubMed]
- Tan W, Zhang W, Strasner A, et al. Tumour-infiltrating regulatory T cells stimulate mammary cancer metastasis through RANKL-RANK signalling. Nature 2011;470:548-53. [Crossref] [PubMed]
- Briseño CG, Satpathy AT, Davidson JT, et al. Notch2-dependent DC2s mediate splenic germinal center responses. Proc Natl Acad Sci U S A 2018;115:10726-31. [Crossref] [PubMed]
- Oh SJ, Ahn S, Jin YH, et al. Notch 1 and Notch 2 synergistically regulate the differentiation and function of invariant NKT cells. J Leukoc Biol 2015;98:781-9. [Crossref] [PubMed]
- Auderset F, Schuster S, Coutaz M, et al. Redundant Notch1 and Notch2 signaling is necessary for IFNγ secretion by T helper 1 cells during infection with Leishmania major. PLoS Pathog 2012;8:e1002560. [Crossref] [PubMed]
- Nakano T, Inoue Y, Shimojo N, et al. Lower levels of hsa-mir-15a, which decreases VEGFA, in the CD4+ T cells of pediatric patients with asthma. J Allergy Clin Immunol 2013;132:1224-7.e12. [Crossref] [PubMed]
- Enomoto A, Ping J, Takahashi M. Girdin, a novel actin-binding protein, and its family of proteins possess versatile functions in the Akt and Wnt signaling pathways. Ann N Y Acad Sci 2006;1086:169-84. [Crossref] [PubMed]
- Trinath J, Holla S, Mahadik K, et al. The WNT signaling pathway contributes to dectin-1-dependent inhibition of Toll-like receptor-induced inflammatory signature. Mol Cell Biol 2014;34:4301-14. [Crossref] [PubMed]
- Silva-García O, Valdez-Alarcón JJ, Baizabal-Aguirre VM. The Wnt/β-catenin signaling pathway controls the inflammatory response in infections caused by pathogenic bacteria. Mediators Inflamm 2014;2014:310183. [Crossref] [PubMed]
- Ye H, Zhang J, Wang J, et al. CD4 T-cell transcriptome analysis reveals aberrant regulation of STAT3 and Wnt signaling pathways in rheumatoid arthritis: evidence from a case-control study. Arthritis Res Ther 2015;17:76. [Crossref] [PubMed]
- Lengfeld JE, Lutz SE, Smith JR, et al. Endothelial Wnt/β-catenin signaling reduces immune cell infiltration in multiple sclerosis. Proc Natl Acad Sci U S A 2017;114:E1168-77. [Crossref] [PubMed]
- Schinzari V, Timperi E, Pecora G, et al. Wnt3a/β-Catenin Signaling Conditions Differentiation of Partially Exhausted T-effector Cells in Human Cancers. Cancer Immunol Res 2018;6:941-52. [Crossref] [PubMed]
- Tang YY, Sheng SY, Lu CG, et al. Effects of Glycogen Synthase Kinase-3β Inhibitor TWS119 on Proliferation and Cytokine Production of TILs From Human Lung Cancer. J Immunother 2018;41:319-28. [PubMed]