
Improving our understanding of breast cancer tumorigenesis across ethnicities
Zhang et al. (1) recently reported in this journal the genetic analysis of 33 breast cancer related genes in breast cancers from patients from the Guangzhou region of China. The study is important since it adds to a growing body of data cataloguing somatic alteration of breast cancer-associated genes in specific ethnic groups (Table 1).
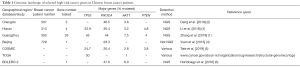
Full table
The study used modern next generation sequencing (NGS) technology to sequence 33 known breast cancer-related genes and compared their findings to Caucasian cases available in The Cancer Genome Atlas (TCGA—excluding the 6% from individuals of Asian ethnicity). It would have been informative for the reader if the authors had compared their data to more than one publicly available resource. The comprehensive cancer gene database, COSMIC (Catalogue of Somatic Mutations in Cancer) (5) would have provided further opportunity for examining ethnic differences in somatic genetic variation.
The authors concluded that there were a number of significant differences in specific gene mutational and copy number profiles between the Chinese and TCGA breast cancer samples. There were some interesting findings such as TP53 and AKT1 pathogenic variants being significantly more common in the Asian cohort. The presence or absence of pathogenic variants themselves was not the only relevant factor uncovered: the genetic profiles within key genes differed between the groups. For example, pathogenic variants in the TP53 tumour suppressor were predominantly missense in the TCGA cohort, whereas other mutation types predominated in the Chinese cohort. As the authors point out, this is immediately significant in that the former mutation types are overall less deleterious to protein function than the latter (7), with clear prognostic implications. It was also of clinical interest that a number of the observed pathogenic variants were actionable/druggable targets. For example, the study found 23 E17K mutations in AKT1.
The authors asked whether the detected genomic alterations were associated with other established clinicopathological variables, in comparison with TCGA data. In summary, 80% of the top 10 most frequently altered genes had statistically different mutation frequencies between the two cohorts, in the hormone receptor-positive/human epidermal growth factor 2-negative patient (HR+/HER2−) group. In contrast, there were no significant differences in mutation frequencies between the two cohorts in the receptor negative (HR−) category. Other studies have examined the association between breast cancer-associated genes and clinicopathological variables in Chinese patients. For example, both Li et al. (3) and Deng et al. (2) found that PIK3CA mutations were statistically more prevalent in HR+ patients (the latter authors also noting that they were also more prevalent in the HER2− group, as were AKT1 pathogenic variants). Deng et al. (2) found no prognostic significance of a single mutation in the PIK3CA/AKT1 pathway, yet two or more mutations were strongly adversely prognostic. One report revealed TP53 pathogenic variants to occur statistically more frequently in ER/PR− cases, and cases with concomitant BRCA1 mutations, a basal histological phenotype or high proliferation as measured by Ki67 immunohistochemistry (3).
However, some critical technical aspects of the study are lacking such that a cautionary caveat should be applied when interpreting this data. The study appears not to have included a sample to represent germline DNA in this analysis; hence, some of the detected variants could have been germline, rather than somatic variants. For instance, there is evidence in individuals of Chinese ethnicity, that pathogenic germline variants in PIK3CA can be found in 5% of the population (2). Other critical aspects of the analysis were equivalent in the two datasets including the technology applied to target the genomic regions of the 33 genes, the sequencing platform and the data filtering pipeline. These are substantial issues in NGS that have been described by many and require considerable attention to study design and time investment to resolve. The authors also acknowledge some drawbacks of their study. It was retrospective, single-institutional and with relatively limited subset size numbers. Additionally, more time is needed for their patient follow-up to mature. Importantly, only 33 genes were tested; it has been estimated that 719 genes have been implicated somatically as cancer drivers (5), and future studies with expanded sample size and gene numbers will further refine our understanding of the frequency of somatic genomic alterations in Asian women with breast cancer and how these may differ from other ethnic populations.
For the future, consistency in technology and data interpretation is critically needed to form a more lucid picture of ethnic differences. Whole genome sequencing might be a good approach, as this is the most comprehensive sequencing format; methods can be standardized and analysis pipelines can be more easily shared. New discoveries will be facilitated by this effort.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Zhang G, Wang Y, Chen B, et al. Characterization of frequently mutated cancer genes in Chinese breast tumors: a comparison of Chinese and TCGA cohorts. Ann Transl Med 2019;7:179. [Crossref] [PubMed]
- Deng L, Zhu X, Sun Y, et al. Prevalence and Prognostic Role of PIK3CA/AKT1 Mutations in Chinese Breast Cancer Patients. Cancer Res Treat 2019;51:128-40. [Crossref] [PubMed]
- Li G, Guo X, Chen M, et al. Prevalence and spectrum of AKT1, PIK3CA, PTEN and TP53 somatic mutations in Chinese breast cancer patients. PLoS One 2018;13:e0203495. [Crossref] [PubMed]
- Yuan H, Chen J, Liu Y, et al. Association of PIK3CA Mutation Status before and after Neoadjuvant Chemotherapy with Response to Chemotherapy in Women with Breast Cancer. Clin Cancer Res 2015;21:4365-72. [Crossref] [PubMed]
- Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019;47:D941-7. [Crossref] [PubMed]
- Hortobagyi GN, Chen D, Piccart M, et al. Correlative Analysis of Genetic Alterations and Everolimus Benefit in Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Advanced Breast Cancer: Results From BOLERO-2. J Clin Oncol 2016;34:419-26. [Crossref] [PubMed]
- Meric-Bernstam F, Frampton GM, Ferrer-Lozano J, et al. Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther 2014;13:1382-9. [Crossref] [PubMed]


