
Implications of the circular RNAs in localized prostate cancer
Stochastic and contingent events in molecular biology tune the advent of new molecular entities, most recent being the circular RNAs (circRNAs) which bestowed new avenues in cancer research. The circRNAs are stable, single-stranded covalently closed transcripts resulting from the back-splicing events of RNA (1). circRNAs are present in the plasma, cell-free saliva and urine of cancer patients and healthy individuals, either in the free forms or bound to the proteins or associated with exosomes highlighting their importance as non-invasive biomarkers (2). The current study by Chen et al. (3) titled, “Widespread and Functional RNA Circularization in Localized Prostate Cancer” has utilized the state-of-the-art-techniques and performed the ultra-deep rRNA depleted non-poly-A RNA sequencing to reveal the circular RNAs profile of localized prostate cancer (PCa), and implicated their role in tumor aggressivity, and specifically cancer cell proliferation, which was unique and independent of other contemporary studies in this field.
Delayed and incorrect detection of cancer is considered as one of the major causes of the cancer related mortality, which necessitates the need to improve upon the sensitive cancer screening techniques for the betterment of patient prognostics. Currently used PCa screening procedures involve digital rectal examination (DRE) and prostate-specific antigen (PSA) level quantification, which lacks the efficiency to distinguish between localized and metastatic stage of the disease, emphasizing that the revelation of new biomarkers to characterize the aggressive clinical and pathologic features of prostate cancer could prove worthwhile. Recently, circular RNAs (circRNAs) have gained attention due to their diagnostic accuracy and applicability as a robust prognostic biomarker of cancer.
Ultra-deep transcriptomic profile of Canadian Prostate Cancer Genome Network (CPC-GENE) cohort
Chen et al. noticed that despite a wealth of genomics and transcriptomics studies in localized PCa, not much was explored about low abundance circular transcripts (circRNA). They aimed to fill this gap by characterizing the transcriptome of 144 localized PCa patients in CPC-GENE cohort (Canadian Prostate Cancer Genome Network), with a median follow-up of 6.5 years which provided rich clinical annotation. To interrogate the unexplored circular RNAs along with linear transcriptome, they performed rRNA-depleted ultra-deep total RNA-sequencing (median 382±138 million reads per tumor) by avoiding poly-A capture. The unsupervised clustering of 4,585 transcripts revealed various subtypes, most of them exhibited high genomic instability, and one showed overlap with the previously defined TCGA-PRAD subtype characterized by CHD1 deletions and SPOP mutations. On comparison of their transcriptomic subtypes with the copy-number alterations (CNA) derived genomic subtypes of localized PCa (4), specific subtype overlaps were found whereas overall concordance was low which suggested that different transcriptional profiles are possible from a single genomic subtype. Further, on identification of the pathways associated with these transcriptomic subtypes, one of the clusters displayed enriched immune-related pathways which highly correlated with tumor-infiltrating immune cells. In addition, they identified 1,223 unique gene fusions with ~14 median fusions per tumor and observed a novel rearrangement between SCHLAP1 lncRNA and UBE2E3 mRNA, which was strongly associated with aggressive disease. Read-through fusion transcripts were present pervasively in their data and pointed towards defective transcription and splicing machinery which suggests towards enhanced circular RNAs biogenesis.
Characterization of the unexplored and low abundance transcripts
To examine the circRNA profiles of different cancer types including prostate, Vo et al. performed the exome capture RNA-seq using RNA probes hybridizing to exons (2), in contrast to the routinely used RNase R (5) and Ribo-Zero techniques (6). Alternatively, the current study utilized the power of ultra-deep rRNA-depleted non-poly-A RNA sequencing with >300× coverage to identify more than hundred thousand transcripts. Here, the authors used the highly sensitive and specific circRNA analysis tool, CIRCexplorer (7) to identify the back-spliced circRNAs, and found 76,311 distinct circRNAs (with at least two back-splicing reads) in CPC-GENE cohort.
While for PCa cell lines, authors performed RNase R treatment followed by RiboMinus RNA-seq, to enrich the circRNAs which are resistant to exonuclease activity. Subsequently, profiles of the patients’ specimens and PCa cell lines were overlapped to generate a list of commonly expressed circRNAs, and about ~25,000 of the circRNAs were nominated. About ~87% of the detected circRNAs belonged to primarily replicating genomic regions suggesting a connection between early replicative genome and circRNAs. The authors deciphered the potential genes which form circRNAs, and found that longer parental genes undergo more splicing events, hence enriched for the regulators involved in their biogenesis. Moreover, only 3% correlation was observed between the abundances of circRNAs and their linear counterparts in patient specimens, highlighting independent mechanisms involved in their biogenesis. They also emphasized the clinical relevance of circRNAs by performing quartile-based classification using circRNA index (CRI), and demonstrated that the patients with enormously high or low circRNAs expression were significantly associated with poor prognosis, which was independent of their linear counterparts. Moreover, higher CRI was positively associated with read-through and fusion events, suggesting a link between fusion and circularization of transcripts. Additionally, they validated these findings using an independent cohort of 49 prostate tumors (NGS-ProToCol), which further emphasized the utility of circRNAs as a prognostic and therapeutic modality. On the contrary, the association of specific circRNAs and linear forms with biochemical recurrence showed a negligible correlation between their hazard ratios. Conclusively, the study accentuates the involvement of distinct biological machineries for generation of circular and linear transcripts.
To explore the functional significance of circRNAs, the authors performed a small hairpin RNA (shRNA)-based loss-of-function screening of the top 2,000 abundant circRNAs in the PCa cell lines followed by analysis through MaGeck, a tool for detecting the essential genes in a RNAi screen, which identified a list of unique 171 circRNAs (8). They next used shRNA screen analysis tool, Achilles shRNA (9), and CRISPR (10) for identification of the context specific genetic dependencies, and found that ~92% of these circRNAs affected cell proliferation independent of their linear counterparts. Further, they validated these results by employing loss-of-function and gain-of-function experiments for randomly chosen 10 circRNAs and observed change in proliferation rate in PCa cell lines. Taken together, authors for the first-time established the role of circular RNAs independent of their linear counterparts in PCa progression.
circCSNK1G3: an apostate to CSNK1G3
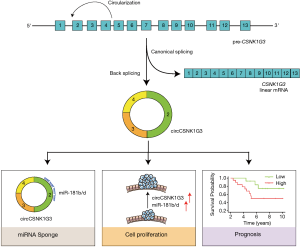
Earlier, the circRNAs were considered a result of defective spliceosomal machinery, however, the recent studies (11) including the one being discussed here, have shown that the production of circRNAs is highly context dependent and selective in nature. Identification of circCSNK1G3 as a transformer of oncogenic properties was in clear contrast to function of its parent gene CSNK1G3; since the authors didn’t see any change in the transcript level of CSNK1G3, while circCSNK1G3 was highly upregulated and was responsible for altering the proliferation rate. The authors have also demonstrated the specific roles of circular and linear forms of CSNK1G3, for instance circCSNK1G3 activated genes were enriched for pathways involved in cell-cycle regulation, whereas CSNK1G3 activated genes were found to be involved in response to acidic proteins, hence, corresponding to the normal function of CSNK1G3. Considering the role of circRNAs as miRNA sponges, the correlation between miRNAs and circCSNK1G3 was examined in the CPC-GENE cohort. Of note, circCSNK1G3 was shown to sequester miR-181b/d, and together they orchestrate the expression of the genes involved in the regulation of cell-cycle, which further indicated that circCSNK1G3 and miR-181b/d governs the PCa cells proliferation (Figure 1).
Future of circRNAs in cancer research
Discovery of circular RNAs could become the next big step in molecular classification of cancer and development of better diagnostics and therapeutics. Recent several studies suggest that the circRNA is the new paradigm shift in the field of cancer, after plethora of studies involving miRNAs and lncRNAs. Vo et al. defined the circular RNA profiles of more than 2,000 cancer patients and established a clinical, cancer-specific database of circRNAs, MiOncoCirc. The discovery of circAURKA in their study can be considered as a surrogate marker for the aggressive neuroendocrine prostate cancer (NEPC), which follows the expression pattern of its parental gene (AURKA) across PCa subtypes (2). On the contrary, the current study by Chen et al. provided evidence that despite having undefined role of linear counterparts in PCa progression, circular RNAs for instance, circCSNK1G3 can play a key role in driving oncogenesis (3). Taken together, these studies are advancing the cancer field by providing opportunities to explore their data for the identification of new target circRNAs, although these as well as other publicly available datasets can be explored for several unknown entities. The enhanced knowledge about circRNAs could furnish the requirement to develop novel biomarkers with high sensitivity and specificity for PCa detection at early stages. Thus, the current study emphasized to think “out of the box” and utilize the available advanced multidisciplinary technologies to their utmost to explore the unexplored.
Acknowledgments
Funding: B Ateeq is supported by the Wellcome Trust/DBT India Alliance grant [IA/I(S)/12/2/500635].
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell 2018;71:428-42. [Crossref] [PubMed]
- Vo JN, Cieslik M, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell 2019;176:869-81.e13. [Crossref] [PubMed]
- Chen S, Huang V, Xu X, et al. Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell 2019;176:831-43.e22. [Crossref] [PubMed]
- Fraser M, Sabelnykova VY, Yamaguchi TN, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017;541:359. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Giannoukos G, Ciulla DM, Huang K, et al. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol 2012;13:R23. [Crossref] [PubMed]
- Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134-47. [Crossref] [PubMed]
- Li W, Xu H, Xiao T, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 2014;15:554. [Crossref] [PubMed]
- Cowley GS, Weir BA, Vazquez F, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data 2014;1:140035. [Crossref] [PubMed]
- Meyers RM, Bryan JG, McFarland JM, et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet 2017;49:1779-84. [Crossref] [PubMed]
- Holdt LM, Kohlmaier A, Teupser D. Circular RNAs as therapeutic agents and targets. Front Physiol 2018;9:1262. [Crossref] [PubMed]


