
Liquid biopsies in renal cell carcinoma with focus on epigenome analysis
We read with great interest the recent paper published by Jung et al. in Clinical Chemistry entitled “Cell-free SHOX2 DNA methylation in blood as a molecular staging parameter for risk stratification in renal cell carcinoma patients: a prospective observational cohort study” (1).
They analyzed “SHOX2 mRNA expression in renal cell carcinoma (RCC) tissues and SHOX2 gene body methylation quantitatively in circulating cell-free DNA (ccfDNA) and RCC tissues with regard to risk stratification”. SHOX2 methylation in formalin-fixed and paraffin-embedded (FFPE) tumor tissue and in the corresponding plasma sample is strongly associated with an advanced tumor stage and risk of death after surgery. They observed that “Pretherapeutic SHOX2 ccfDNA methylation testing allows for the identification of RCC patients at high risk of death after nephrectomy. These patients might benefit from an adjuvant treatment or early initiation of a palliative treatment”. The prognostic value of SHOX2 methylation was evidenced in both tissue and plasma samples analysis. Hypermethylated SHOX2 patients experienced a worse overall survival compared to the hypomethylated, according to an optimized methylation cutoff (19.11%), established with regard to the greatest survival difference between methylation-negative and -positive patients (1).
SHOX2 and its role in tumorigenesis
The results presented by Jung et al. pave the way to numerous observations and comments. First of all, as also discussed by Jung and colleagues, the role of SHOX2 during tumorigenesis could be part of epithelial-to-mesenchymal transition (EMT) process via transforming growth factor β signaling (2,3). EMT is frequently observed in clear cell RCCs as the results of a sarcomatoid differentiation. A sarcomatoid phenotype in RCC has to be considered as grade 4 as reported in the WHO 2016 classification and it is positively associated with advanced stage and poor prognosis (4).
At single-cell level, EMT consists in a morphological transformation of RCC cells that leads to the loss of surface epithelial antigens and to the acquisition of mesenchymal features (i.e., vimentin expression) (5,6).
Liquid biopsy (LB) in RCC has received great attention for its potential prognostic and predictive value in helping clinicians to better understand the biology of the tumor and to personalize treatment in a non-invasive way (7). One of the most important limits in the application of LB in RCC is the low sensitivity in the detection of circulating tumor cells (CTC) by Epithelial Marker-dependent isolation methods. Indeed, the loss of surface epithelial antigens during the EMT process prevent the possibility to capture CTCs. The implementation of other surface markers such as antibodies directed against membrane Carbonic anhydrase 9 (CA9/CAIX) and CD147 allows the isolation of a greater number of CTCs (8).
DNA methylation changes have been linked with stemness and metastasis in circulating tumor microemboli (CTMs). CTM, another promising entities detectable with LB, are defined as a group of cell, from two to more than 50 CTCs, mixed with leukocytes, cancer-associated fibroblasts, endothelial cells, and platelets (9,10). The higher metastatic potential and their property to remain “dormant” for long period have been ascribed to a remarkable enrichment for stemness-related transcription factors that coordinately regulate proliferation and pluripotency. Differently, single CTCs featured hypomethylation of other transcription factor binding sites, including those that are occupied by MEF2C, JUN, MIXL1, and SHOX2, commonly enriched in various cancers (11).
Epigenome studies in liquid biopsy and genitourinary tumors
The current ccfDNA detection methods have demonstrated to be more efficient in the advanced stage setting rather than in cancer screening and the detection of minimal residual disease after treatment. Nonetheless, the presence of ccfDNA is not specific for a tumor condition. Indeed, high levels of ccfDNA have been found also in patients with acute blunt trauma, burn victimizes, sepsis, and myocardial infarction (12-14). ccfDNA quantification does not allow to discriminate which DNA fragments derived from cancer cells (circulating tumor DNA—ctDNA) or from a necrotic inflammatory process. In order to distinguish ctDNA from ccfDNA, novel detection strategies have been developed.
Fleischhacker et al. settled a new sensitive, immunoprecipitation-based protocol to explore the methylome of low quantities of ccfDNA. Cancer-specific differentially methylated regions (DMRs) allow ctDNA detection with high sensitivity, low-cost, and high-efficiency method (15). CpG island hypermethylation of ccfDNA in patients with RCC has been investigated as potential diagnostic biomarker by other authors. Hypermethylation on ccfDNA was found for the LRRC3B (74.1%), APC (51.9%), FHIT (55.6%), and RASSF1 (62.9%) genes in RCC patients (16).
Hauser et al. also demonstrated a higher methylation frequency in RCC patients compared to healthy individuals. The sensitivity of the methylation assay was low in single-gene analysis, but combined analysis of methylation frequency of multiple genes (i.e., APC, PTGS2 and GSTP1) reached a sensitivity of 62.9% and a specificity of 87%. DNA hypermethylation of APC gene was associated with advanced cancer stage (17).
Other genes such as SFRP1, BNC1, GREM1, RASSF1A, PCDH8, SCUBE3, GATA5, LAD1 and NEFH, promoter methylation has found to be associated with patient outcome, and their prognostic value was independently validated (18-20). Superior prognostic value has been obtained by combination of several markers as compared to the markers alone. Four-marker panel based on methylation level of GREM1, LAD1, NEFH and NEURL has proved to be able to identify patients with poorer survival in two independent patient series (21). The prognostic risk score based on a five-CpG-based-classifier developed by Wei and colleagues may be adopted to separate patients within the same clinical stage into subgroups with better and worse prognosis (22).
A diagnostic urine essay based on DNA methylation of OTX1, ONECUT2 and TWIST1 combined with mutation analysis in FGFR3, TERT and HRAS has been validated in patients with hematuria prior to cystoscopy. The area under the curve (AUC) obtained was of 0.96 with 93% sensitivity and 86% specificity and an overall negative predictive value of 99%. An appropriate selection of patients candidate to cystoscopy by this predictive essay may lead to a reduction of costs and overtesting (23). Another important result has been obtained by the use of epigenetic essay in prostate cancer patients candidate to a re-biopsy after a prior negative biopsy. The methylation ratio of 3 genes GSTP1, APC and RASSF1 were assessed in the FFPE from the initial biopsy. A negative predictive value of 88% was reached and the essay has proved to be an independent predictor of prostate cancer detection in a repeat biopsy (24).
ccfDNA compared to other liquid biopsy’ entities
Compared with CTCs and CTMs, ccfDNA represents a better biomarker in terms of feasibility and reproducibility. Its half-life is less than 2 h and it is more stable than cells; ccfDNA is more sensitive than CTC Assay, and easily detectable. From a biological point of view, ctDNA is better biomarker in monitoring tumor dynamics showing a greater correlation with changes in tumor burden. However, ctDNA in most cases requires a priori knowledge of the gene of interest and not all DNA mutations can be found. Whole exome sequencing (WES), splice variants, transcriptome analyses and functional assays can be performed only with CTC (25).
DNA methylation is a stable epigenetic mark and its quantification can be performed in both solid biopsy (i.e., FFPE tissue) and LB (i.e., blood, stool, and urine). Consequently, methylation analysis has considered a potential diagnostic, prognostic, and predictive biomarkers (26,27) (Figure 1).
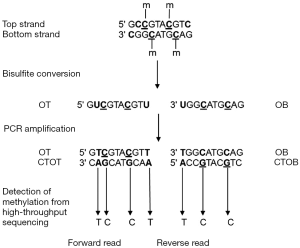
Future perspectives and conclusions
Further studies are urgently needed in order to select specific genes of interest, or gene panels, and also to determine a validated optimized cut-off to better discriminate patients with higher risk of recurrence and metastatic progression. RCC may absolutely benefit from the development of non-invasive and reliable biomarkers, allowing early and timely personalized treatment changes. Many efforts for implementation of liquid biopsies are currently under examination along with emerging liquid biopsy entities (i.e., EVs, ccfRNAs, branched- chain amino acids (BCAAs), proteins, tumor- educated platelets).
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Jung M, Ellinger J, Gevensleben H, et al. Cell-free SHOX2 DNA methylation in blood as a molecular staging parameter for risk stratification in renal cell carcinoma patients: a prospective observational cohort study. Clin Chem 2019;65:559-68. [Crossref] [PubMed]
- Hong S, Noh H, Teng Y, et al. SHOX2 is a direct miR-375 target and a novel epithelial-to-mesenchymal transition inducer in breast cancer cells. Neoplasia 2014;16:279-90.e1-5.
- Yi J, Jin L, Chen J, et al. MiR-375 suppresses invasion and metastasis by direct targeting of SHOX2 in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) 2017;49:159-69. [PubMed]
- Moch H, Cubilla AL, Humphrey PA, et al. The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: renal, penile, and testicular tumours. Eur Urol 2016;70:93-105. [Crossref] [PubMed]
- Montironi R, Santoni M, Scarpelli M, et al. Re: epithelial-to-mesenchymal transition in renal neoplasms. Eur Urol 2015;68:736-7. [Crossref] [PubMed]
- Piva F, Giulietti M, Santoni M, et al. Epithelial to Mesenchymal transition in renal cell carcinoma: implications for cancer therapy. Mol Diagn Ther 2016;20:111-7. [Crossref] [PubMed]
- Cimadamore A, Gasparrini S, Massari F, et al. Emerging molecular technologies in renal cell carcinoma: liquid biopsy. Cancers (Basel) 2019;11. [Crossref] [PubMed]
- Liu S, Tian Z, Zhang L, et al. Combined cell surface carbonic anhydrase 9 and CD147 antigens enable high-efficiency capture of circulating tumor cells in clear cell renal cell carcinoma patients. Oncotarget 2016;7:59877-91. [PubMed]
- Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. [Crossref] [PubMed]
- Zhang D, Zhao L, Zhou P, et al. Circulating tumor microemboli (CTM) and vimentin+ circulating tumor cells (CTCs) detected by a size-based platform predict worse prognosis in advanced colorectal cancer patients during chemotherapy. Cancer Cell Int 2017;17:6. [Crossref] [PubMed]
- Gkountela S, Castro-Giner F, Szczerba BM, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 2019;176:98-112.e14. [Crossref] [PubMed]
- Rhodes A, Wort SJ, Thomas H, et al. Plasma DNA concentration as a predictor of mortality and sepsis in critically ill patients. Crit Care 2006;10:R60. [Crossref] [PubMed]
- Chang CP, Chia RH, Wu TL, et al. Elevated cell-free serum DNA detected in patients with myocardial infarction. Clin Chim Acta 2003;327:95-101. [Crossref] [PubMed]
- Lu H, Busch J, Jung M, et al. Diagnostic and prognostic potential of circulating cell-free genomic and mitochondrial DNA fragments in clear cell renal cell carcinoma patients. Clin Chim Acta 2016;452:109-19. [Crossref] [PubMed]
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer—a survey. Biochim Biophys Acta 2007;1775:181-232. [PubMed]
- Skrypkina I, Tsyba L, Onyshchenko K, et al. Concentration and methylation of cell-free DNA from blood plasma as diagnostic markers of renal cancer. Dis Markers 2016;2016:3693096. [Crossref] [PubMed]
- Hauser S, Zahalka T, Fechner G, et al. Serum DNA hypermethylation in patients with kidney cancer: results of a prospective study. Anticancer Res 2013;33:4651-6. [PubMed]
- Ricketts CJ, Hill VK, Linehan WM. Tumor-specific hypermethylation of epigenetic biomarkers, including SFRP1, predicts for poorer survival in patients from the TCGA Kidney Renal Clear Cell Carcinoma (KIRC) project. PLoS One 2014;9:e85621. [Crossref] [PubMed]
- Morris MR, Ricketts C, Gentle D, et al. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene 2010;29:2104-17. [Crossref] [PubMed]
- Morris MR, Ricketts CJ, Gentle D, et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene 2011;30:1390-401. [Crossref] [PubMed]
- van Vlodrop IJH, Joosten SC, De Meyer T, et al. A four-gene promoter methylation marker panel consisting of GREM1, NEURL, LAD1, and NEFH predicts survival of clear cell renal cell cancer patients. Clin Cancer Res 2017;23:2006-18. [Crossref] [PubMed]
- Wei JH, Haddad A, Wu KJ, et al. A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nat Commun 2015;6:8699. [Crossref] [PubMed]
- van Kessel KE, Beukers W, Lurkin I, et al. Validation of a DNA methylation-mutation urine assay to select patients with hematuria for cystoscopy. J Urol 2017;197:590-5. [Crossref] [PubMed]
- Partin AW, Van Neste L, Klein EA, et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol 2014;192:1081-7. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Schröck A, Leisse A, de Vos L, et al. Free-circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: an observational prospective cohort study. Clin Chem 2017;63:1288-96. [Crossref] [PubMed]
- David SA, Mersch M, Foissac S, et al. Genome-wide epigenetic studies in chicken: a review. Epigenomes 2017;1:20. [Crossref]


