
TSPAN8 serves as a prognostic marker involving Akt/MAPK pathway in nasopharyngeal carcinoma
Introduction
Although Nasopharyngeal carcinoma (NPC) is not common, it ranks out of ten common malignancy and 10th frequent cause of cancer deaths. The incidence rates are low throughout most of the world, usually 1 per 100,000 population, but it is more common in southern China (1) and its incidences vary by geographic location (2). Genetic susceptibility and EBV infection are two main risk factors for NPC development (3-6). Despite the development of worldwide prevention efforts, incidences of NPC are increasing by the day. Radiotherapy is the main treatment for NPC. However, tumor relapse and distant metastases remain major obstacles for the long-term survival of NPC patients (7,8). Although several studies have improved our general understanding of the NPC pathogenesis, none of the studies focused on elucidating NPC specific biomarkers. Therefore, it is essential to carry out an in-depth study of the molecular mechanisms of NPC to find the appropriate biomarkers for NPC diagnosis and therapy.
Previous reports have shown that genetic changes were frequently associated with the development of NPC, such as the specific HLA types (7), polymorphisms in CYP2E1 (9) and P450 2E1 (10), Wnt signaling pathway protein, and hOGG1 and XRCC1 involved in DNA repair (11). Gene expression profile is a high throughput and rapid method used to detect the gene expression level. Several studies have used microarray for genome scale analysis in clinical specimens and cell lines, especially to identify the genes that contribute to the NPC genesis (12,13). Zeng et al. analyzed the gene expression profiles in 23 NPCs and 10 normal nasopharyngeal epithelial (NPE) tissue samples; and found the alterations in the WNT pathway suggesting that this pathway may be activated in NPC. The data generated in this study presented a comprehensive list of genes aberrantly regulated in NPC, for example; SYTL2, AGTRL1 and RAPGEF5 (12). In another study, Fung et al. had examined the differential gene expression in non-malignant and malignant NPE cells using a cDNA array hybridization method, and found that calgranulin A, calgranulin B, ENA-78, FRA-1 and NF90 were highly expressed in malignant NPE cells. Several studies had found genes identified in NPC, which was reported to be markers of metastasis such as serine protease inhibitor Kazal-type 6 (SPINK6), but most of these studies did not elucidate the molecular mechanism involved in the metastasis of NPC. Therefore, it is of vital importance to identify a gene that relate to the metastatic pathway of NPC in order to generate a new prognostic marker for the detection of NPC progression.
In this study, we identified a gene, TSPAN8 and confirmed its overexpression in NPC tissues and poorly differentiated and highly-metastatic cell lines. TSPAN8 is a member of the tetraspanins of integral membrane protein, and is characterized by four highly conserved transmembrane domains which is also known as transmembrane 4 superfamily 3 (TM4SF3). TSPAN8 have emerged as an important player in cancer (14,15), immune disease (16), and infectious disease (17,18). There has been many studies elucidating the role of TSPAN8 in tumor progression and angiogenesis (19-21), and is found to be significantly increased in pancreatic adenocarcinoma (22). Moreover, higher expression of TSPAN8 are associated with poor prognosis in esophageal carcinoma (23,24), melanoma (25) and gastric carcinoma (26). TSPAN8 might promote the progression and metastasis of cancer cell by increasing cellular movement and deregulating adhesion (27,28). However, little is known about the role of TSPAN8 in NPC.
This study sought to identify new biomarkers associated with NPC progression. Microarrays were used to identify genes that varied in expression level between primary NPC and corresponding benign adjacent tissues. Among them, TSPAN8 was highly expressed in NPC tissue and the expression of TSPAN8 is associated with poor survival in NPC patients. TSPAN8 was highly expressed in NPC cell lines is involved in the Akt/MAPK signaling pathway. Importantly, our results confirmed that TSPAN8 was an important biomarker of NPC progression and metastasis. To the best of our knowledge, this is the first report to show that TSPAN8 could be a prognostic factor and therapeutic target for NPC patients.
Methods
Patient and tissue specimens
The patients enrolled into this study were diagnosed with NPC at the Department of Nasopharyngeal Carcinoma, Sun Yat-sen Cancer Centre. A total of 104 paraffin embedded NPC samples were examined. the written informed consent was obtained from all patients, and the protocol was approved by the Research Ethics Board at Sun Yat-sen Cancer Centre. NPC specimens were staged in accordance with the UICC classification guidelines. A long-term follow-up of 104 cases of nasopharyngeal squamous cell carcinoma was done 80 months after the first occurrence; and the local recurrence, distant metastasis and survival time were recorded. NPC patient’s data analyses were performed via survival analyses. One hundred and four cases of NPC with sufficient follow-up data were qualified for analyses after immunohistochemistry (IHC) staining for TSPAN8.
mRNA expression profiling
To carry out mRNA expression profiling, Arraystar Human mRNA Expression Profiling Service Report based on polymerase chain reaction (PCR) technique was used (KANGCHEN, Shanghai, China). Agilent Array platform was employed for four pairs of tumor tissues and their corresponding benign adjacent tissues from the NPC patients. The sample preparation and microarray hybridization were performed based on the manufacturer’s standard protocols with minor modifications. Functional annotations, such as gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathways, were analyzed using database for annotation, visualization and integrated discovery.
Cell lines
NPC cell lines with various differentiation and metastatic ability were used in this study. CNE1, highly differentiated; CNE2, poorly differentiated, subclone S18, high metastatic ability; subclone S26, low metastasis ability; and NP69, immortalized nasopharyngeal epithelial cell were obtained from Department of Nasopharyngeal Carcinoma, Sun Yat-sen cancer Centre. These cancer cells were maintained in RPMI-1640 supplemented with 10% Fetal Bovine Serum, 5% CO2 at 37 °C; immortalized nasopharyngeal epithelial cell NP69 were maintained in Keratinocyte Serum Free Medium with Epidermal Growth Factor and Bovine Pituitary Extract (Invitrogen Life Technologies, Carlsbad, CA, USA.) The NPC line CNE-2 and its subclones S18, and S26 have previously been established and reported (29).
Plasmid construction and transfection
The full-length TSPAN8 cDNA (GenBank accession number NM_004616.2) was cloned into the GV143 expression vector (Genechem, Shanghai, China). The NPC cells were cultured to 80% confluence in a 6-well plate for 24 h before transfection. The cells were transfected with GV143 and GV143/TSPAN8 plasmid using Lipofectamine™ 2000 according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were collected to perform subsequent assays.
TSPAN8 siRNA (siTSPAN8) transfection
Three sequences of TSPAN siRNAs were designed by Genepharma (Guangzhou, China). S18 and CNE2 cells were grown to approximately 60% confluence in antibiotic-free growth medium and transfected with the three siRNAs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Cells were transfected with 50 nM TSPAN8 siRNA or control. Cells were harvested 24 and 48 h after siRNA transfection and knockdown efficacy was assessed by real time-polymerase chain reaction (RT-PCR) and western-blot. After transfection, cells were detached used for RT-PCR, western blot, migration and invasion assays.Among these siRNAs, two siRNAs were determined to have high gene silencing efficacy (75–81%).
Real time quantitative PCR
Total RNA from specimens or cell lines was extracted with the Trizol reagent (Invitrogen Life Technologies, CA, USA). cDNA was obtained by reverse transcription of the total 500 ng RNA with a Reverse Transcription Kit (Takara Inc, USA). GAPDH was used as an internal control, and gene values were normalized to those of GAPDH. Real-time PCR was performed on a Roche LightCycler480 Real-Time PCR System. All experiments were done in triplicate.
Western-blot
Cells were treated with either DMSO or AKT inhibitor MK2206 (Selleck, TX, USA) for 24 h after transfection. Cell lysates were prepared with RIPA lysis solution. Protein concentration was measured using the BCA protein assay kit (PIERCE, Rockford, IL). Equal amount of protein was run on 12% SDS-PAGE gel and the proteins were electro-transferred from the gel to PVDF membrane. The membrane was then blocked with 5% non-fat milk solution for 1 h, incubated with primary monoclonal antibody against TSPAN8 (1:500, R&D, USA) and Akt, p-Akt, P38, p-P38, ERK, p-ERK (1:1,000, CST, USA) at 4oC overnight. Anti-GAPDH (1:1,000, Santa Cruz, CA, USA) was used as an internal control. The membrane was subjected to immunoblotting using HRP-conjugated antibody. The membrane was detected by enhanced chemiluminescence (ECL) detection system.
Colony formation assay
Five hundred cells were plated in 6-well plate and cultured with RPMI 1640 supplemented with 10% FBS for 10–14 days until colonies were visible. The colonies were fixed with 4% formaldehyde for 15 min and stained with 1% crystal violet for 1 min. Colony were only counted if colony size reached 10 cells cluster in microscopy. All experiments were done in triplicate.
Migration and invasion assay
Briefly, 1×105 cells in (RPMI1640/ 0.2% bovine serum albumin) were added to the upper chambers with 8 mm pore filter of the inserts of a 24-well culture plate (Corning, NY,USA) while RPMI1640/10% FBS were added to the lower chambers. After 8 h, the non-migrated cells were gently removed from the upper chamber with cotton swabs. The cells that had passed through the filters to the lower sides of the chambers were stained with crystal violet and photographed.
ELISA assay
CNE2 cells were cultured in RPMI-1640/10% FBS, CNE2 cells were transiently transfected by TSPAN8 SiRNA for 48h and the IL-1βexpression level was detected with ELISA. Cell supernatant were collected, 3,000 r, 10 min at 4 °C. One hundred µL 1× capture antibody was added in 96-well culture plate at 4 °C overnight, after Wash Buffer washing, 1× ELISA/ELISPOT Diluent was added in every well. 20 µL of each prepared Standard dilution was added. The sample wells were added with the cell supernatant in duplicate. An adhesive film was Covered and incubated at room temperature for 2 h. Biotin-Conjugate prepared. all wells were added with Biotin-Conjugate. Adhesive film was removed and wells were emptied. All wells were added with diluted Streptavidin-HRP, including the blank wells. Adhesive film was covered and incubated at room temperature. The absorbance value of each well was measured with a microplate reader set. Each experiment was repeated three times.
IHC
All 104 specimens of NPC patients were paraffin-embedded. After dewaxing and hydration, the slides were washed with PBS and endogenous peroxidase activity was blocked with 3% hydrogen peroxide. After heating at autoclave for 10 min, 1% hydrogen peroxidase was used to block endogenous peroxidase activity. The specimens were then incubated with anti-TSPAN8 (1:50, R&D, USA) for 90 min at room temperature. After PBS washing, the samples were incubated with secondary antibodies for 60 min at room temperature. Finally, DAB staining was performed, followed by counterstaining of with hematoxylin. The stainings were evaluated by calculating the average positively stained tumor cells in high magnification using an inverted optical microscope by two independent reviewers who were blinded to the clinical parameters.
Statistical analysis
All statistical analyses were performed using SPSS 13.0 and GraphPad 5.0 software. Data were presented as mean ± SD. T-test was used to compare two independent groups of data. The median IHC staining score via Allred score was used as the cutoff value to divide the patients into low and high TSPAN8 expression groups. The χ2 test for proportion and Pearson’s correlation test were used to analyze the relationship between TSPAN8 expression and clinicopathologic characteristics. Kaplan-Meier survival curves were plotted, and log-rank tests were performed. The significance of several variables for survival was analyzed using the Cox regression model in a multivariate analysis. A P value less than 0.05 was considered statistically significant.
Results
mRNA expression profiles of NPC patients and the validation of candidate genes
To identify the altered mRNAs in NPC tissues, mRNA profiling based on Differential Display of reverse Transcriptional PCR were performed using Arraystar Human mRNA Expression Profiling Service Report from Kangchen (Shanghai, China). Among the four pairs of tumor tissues and their corresponding benign adjacent tissues from NPC patients, 1,787 genes were differentially expressed (minimally 2-fold difference), with 199 genes being upregulated more than 10 folds, and 90 genes being upregulated more than 25 folds in NPC patient samples. Among them, these 8 genes were among the most upregulated expressed (Figure 1A), namely: were prolactin-induced protein (PIP), secretoglobin family 1A (SCGB1A1), BPI fold containing family A member 1 (PLUNC), secretoglobin family 2A (SCGB2A1), tubulin polymerization promoting protein (TPPP), transmembrane 4 superfamily 3 (TSPAN8), chemokine C-X-C motif ligand 17 (CXCL17), A kinase PRKA anchor protein 14 (AKAP14), secretory leukocyte peptidase inhibitor (SLPI), rabaptin, RAB GTPase binding effector protein 2 (RABEP2), CREB3 (cAMP responsive element binding protein 3) and cytochrome P450, family 4, subfamily x, polypeptide 1 (CYP4X1). These genes were selected for validation by qRT-PCR. Among these 8 genes, only TSPAN8 was consistently highly expressed in both CNE2 and S18 cell lines, therefore is a promising candidate as a prognostic biomarker for NPC (Figure 1B).
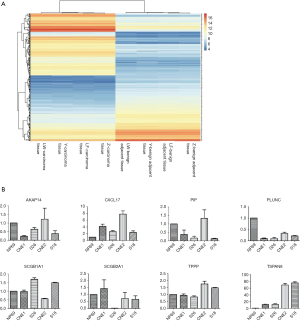
TSPAN8 is highly expressed in NPC tumor tissues and various NPC cell lines
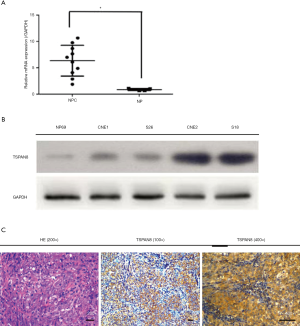
To determine the expression of TSPAN8 in NPC cell lines, qRT-PCR, IHC and Western-blot were performed in CNE1, CNE2, S26, S18 cell lines. TSPAN8 mRNA expression level was determined in ten NPC tissues and five corresponding benign adjacent tissues (Figure 2A). TSPAN8 mRNA expression in NPC tumors was significantly higher (P<0.05) than the benign adjacent tissues (Figure 2A). In vitro, TSPAN8 protein level was especially overexpressed in highly-invasive NPC cell lines, S18 and CNE2 (Figure 2B) and IHC staining revealed that TSPAN8 expression was localized in the cancer cell membrane as shown in Figure 2C.
TSPAN8 silencing in NPC cells inhibited cell migration and invasion
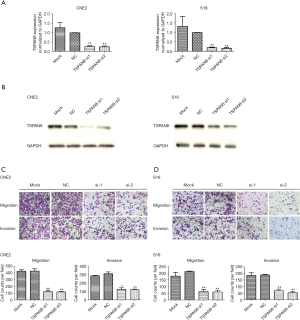
To examine the functions of TSPAN8 in NPC, siRNA was used to silence TSPAN8 in S18 and CNE2 cells. After the cells were transfected with either TSPAN8 siRNA or control siRNA for 24 h, the cells were collected for RT-PCR analysis. As seen in Figure 3A, both TSPAN8 mRNA and protein level were significantly decreased in CNE2 and S18 cells after TSPAN8 silencing (Figure 3A,B). The migration and invasion activities of CNE2 and S18 cells were compared after transfection with TSPAN8 siRNA or control siRNA using transwell assay (Figure 3C,D). These results suggest that depletion of TSPAN8 can inhibit migration and invasion of NPC cell.
Overexpression of TSPAN8 in NPC cells promoted cell migration and invasion
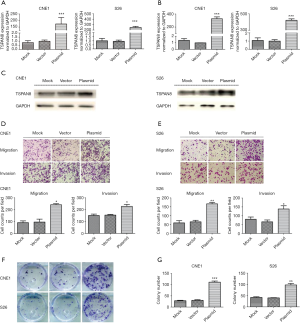
To further determine the role of TSPAN8 in tumor progression, TASPAN8-CMV-MCS-EGFP-SV40-Neomycin vector was transiently transfected into the low-invasive NPC CNE1 and S26cell lines, which were also identified as low TSPAN8 expressing cells. After optimization, plasmid concentration of 1.5 µg/mL were selected for subsequent experiment (Figure 4A). As demonstrated by RT-PCR and Western Blot, the mRNA and protein level of TSPAN8 was significantly increased in both CNE1 and S26 cells after TSPAN8 vector transfection (Figure 4B,C). Migration and invasion of CNE1 cells (Figure 4D), and S26 cells (Figure 4E). Importantly, the overexpression of TSPAN8 significantly promoted colony formation (Figure 4F,G).
Akt/MAPK pathway is involved in TSPAN8 mediated metastasis
To explore the molecules related to NPC metastasis pathway, TSPAN8 were silenced in CNE2 cell using TSPAN8 siRNA. From the Tumor Metastasis Pathway Finder PCR array, specific knockdown of TSPAN8 led to the down-regulation of IL-1β, CTBP1, MMP10, MMP7, PLAUR, IGFBP3, SERPINB2 and SNAIL1 (Figure 5A). Among them, IL-1β showed the most downregulation (6.98 folds). siRNA silencing of TSPAN8 also significantly altered the level of IL-1β protein (Figure 5B). Since IL-1β is a regulating factor of the Akt/MAPK pathway, the effect of TSPAN8 silencing on this pathway was elucidated. As seen in Figure 5C, TSPAN8 silencing decreased the phosphorylation of Akt, ERK and P38, suggesting a role for TSPAN8 as a mediator in the Akt/MAPK pathway.
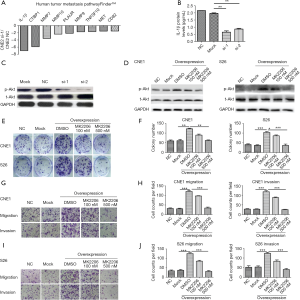
To further verify the role of TSPAN8 in Akt pathway, we used Akt inhibitor MK2206 to rescue the effect induced by overexpressed of TASPAN8. After the transfection of (TASPAN8-CMV-MCS-EGFP-SV40-Neomycin) to CNE1 and S26 cells, p-Akt was significantly overexpressed compared to the lipofectamine-transfected group (Mock) and the empty plasmid group (Vector). At 100 nM, Akt inhibitor MK2206 partially down-regulate induced due to overexpression of TASPAN8 at 100 nM and further down-regulation of p-Akt was seen at 500 nM of MK2206 in CNE2 and S26 (Figure 5D). MK2206 also down-regulated colony formation, migration and invasion in CNE2 and S26 (Figure 5E,F,G,H,I,J).
Correlation between TSPAN8 and clinicopathological features and survival in NPC patients
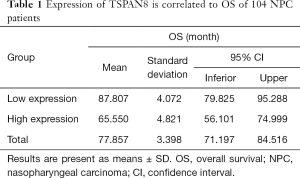
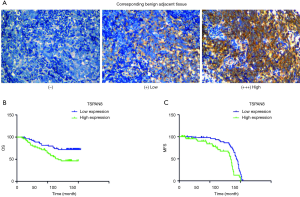
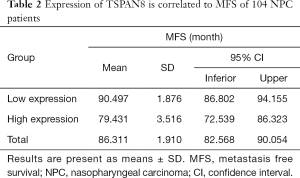
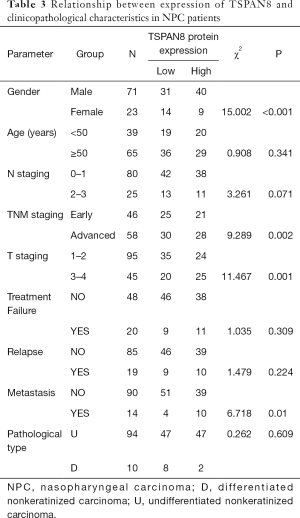
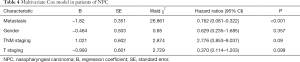
NPC tumors exhibited high expression of TSPAN8 protein, significantly higher than that in normal nasopharyngeal epithelium. We next analyzed the correlation between the expression of TSPAN8 protein and clinicopathological characteristics of NPC. As summarized in Table 1, the expression of TSPAN8 was independent of gender, age, pathology classification, N classification, T classification and TNM classification (the most commonly used tumor staging system in the world at present). The long-term follow-up data indicated that there was a significant relationship between TSPAN8 and metastasis, but not with the disease relapse. TSPAN8 overexpression was positively associated with NPC patients compared with nasopharyngeal non-carcinoma patients (Figure 6A). To evaluate the association between TSPAN8 expression and patient’s outcome, Kaplan–Meier curves were plotted. As shown in Figure 6B,C and Tables 1-4, high TSPAN8 expression led to a shorter overall survival and metastasis free survival (MFS) compared to those with low TSPAN8 expression.
Full table
Full table
Full table
Full table
Discussion
Compared with other cancer types, NPC is uncommon, albeit with a very unique pattern of geographical distribution, and most patients did not high survival rate due to tumor relapse and metastasis. Therefore, urgent efforts are needed to identify reliable tumor markers for early detection and to search for NPC-specific cellular targets for novel therapeutic approaches.
Using PCR technique via mRNA expression profiling with four pairs of tumor tissues and their corresponding benign adjacent tissues from NPC patients, we found that 1,787 genes were differentially expressed (minimally 2-fold difference), Within these genes, only TSPAN8 was consistently highly expressed in both poorly differentiated CNE2 and highly-metastatic subclone S18 cell lines. Claas et al., reported that TSPAN8 exerts a wide spectrum of physiological activities, such as inflammation, tumor progression, and consumption coagulopathy (30). Overexpression of TSPAN8 enhanced metastasis, promotes invasion and migration in several kinds of carcinomas, such as esophageal carcinoma (23), hepatocellular carcinomas (30), pancreatic adenocarcinoma (22) and colorectal carcinomas (27,31).
The current study found that TSPAN8 promote invasion and migration activities in NPC cell lines and tissue via Akt/MAPK signaling pathways. TSPAN8 mRNA and protein levels were found to be elevated in human NPC tissues compared to normal tissues. IHC analysis indicated that TSPAN8 levels were increased in metastatic tumors, compared with those early-stage tumors. And higher TSPAN8 expression level was associated with higher histological differentiation and worse overall survival. Knockdown of TSPAN8 by siRNA transfection resulted in significant inhibition of NPC migration and invasion in vitro while the overexpression of TSPAN8 showed the opposite effects. In addition, TSPAN8 promoted cancer cell migration by regulating the expression of relevant genes, such as ADAM12m. We found that activation of the MAPK pathway was involved in the metastatic functions of TSPAN8 in NPC. To the best of our knowledge, our study is the first demonstration of TSPAN8 exerted to promote invasion activities in NPC cell lines and tissue via Akt/MAPK signaling pathways.
TSPAN8 is a transmembrane protein that is abundantly expressed, consists of small and large outer loops, short N-terminal and C-terminal tails, and four transmembrane domains (32-34). Interaction of TSPAN8 with integrin induces it to a migration supporting manner to increase tumor cell migration (19). Guo and colleagues identified that TSPAN8-associated cell migration was accompanied by the upregulation of both integrin-dependent cell matrix adhesion and calcium-dependent cell-cell adhesion. TSPAN8 functions as a regulator of both cell matrix and cell-cell adhesion (27). Zhou et al. reported ADAM12m is a key matrix degradation enzyme involved in TSPAN8 promoted metastasis. Upregulation of TSPAN8 inhibited MMP2 activation in five stable overexpressed clones in 3D collagen I culture (23). Recent study showed that tumor exosome which constitute TSPAN8 and CD151 promoted matrix degradation, reprogram stroma and drive non-metastatic cells to a motile phenotype (35).
Moreover, we found that the knockdown of TSPAN8 led to the down-regulation of several genes related to Akt/MAPK pathway, and among them, IL-1β showed the most significant downregulation. IL-1β is a pro-inflammatory gene and has been identified to mediate a critical role in suppression of apoptosis, proliferation, angiogenesis, invasion, and metastasis (36). Autocrine production of interleukin IL-1β promotes growth in pancreatic carcinoma cell lines (37). IL-1β secretion into the tumor milieu also induces several angiogenic factors from tumor and stromal cells that promotes tumor growth through hyper neovascularization in lung carcinoma growth in vivo (38). Al-toub et al. identified tumor-derived IL-1β as the prominent cytokine responsible for induction of inflammatory response in MSCs and signaling via mitogen activated protein kinase kinase (MAPKK), contributing to tumorigenicity (39). The overexpression of IL-1β in turn promotes glioma growth, induces activation of the p38 MAPK pathway, increases the GSC phenotype, and upregulates the cancer stem cell phenotype in murine and human Proneural glioma stem cells (GSCs) (40). Wang et al. found that IL-1β regulated the expression of glucocorticoid receptor isoforms in nasal polyps in vitro via p38 MAPK and JNK signaling transduction pathways (41). Based on the literatures, we intend to elucidate the involvement of the Akt/MAPK pathway in the metastatic functions of TSPAN8 in NPC. Our study is the first demonstration of TSPAN8 exerted to promote invasion activities in NPC cell lines by cell function assay and may play an important role in Akt/MAPK signaling pathways.
In addition, MAPK and Akt signaling pathways activation can lead to tumor cell migration and invasion via different mechanisms. The most extensively studied groups of MAPKs are the ERK1/2, JNKs and P38 isoforms, which appear to have independent functions (42). They have been implicated in a wide range of complex biologic processes, such as cell proliferation, differentiation, death, migration and invasion. Therefore, MAPK pathway plays an important role in cancer metastasis. The activation of Akt signaling is a prominent pathway in metastatic breast cancer cells, ovarian cancer cells and lung cancer cells (43-45). Akt signaling pathway was found to be involved in promoting cancer cell metastasis by degradation of extracellular matrix (46).
Conclusions
In summary, the present study suggested that TSPAN8 may play a role in tumor progression and metastasis through Akt/MAPK pathway in NPC. TSPAN8 acts as a tumor migration marker and may be a prognostic factor or therapeutic target for NPC patients, which could be considered a target for NPC therapy.
Acknowledgements
Funding: This work was supported by grants from the National Natural Science Foundation of China (Grant No.: 81672680, 81472525,81372819, 81572596, and U1601223); Natural Science Foundation of Guangdong Province (Grant No.: 2017A030313828); Guangzhou Science and Technology Program (Grant No.: 201704020131).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The work was performed in accordance with the Institutional Review Boards of Sun Yat-sen University Cancer Center (GZR2016-210). Written informed consent was obtained from each patient, including signed consent for tissue analysis and consent to be recorded for potential medical research at the time of sample acquisition.
References
- Jia WH, Feng BJ, Xu ZL, et al. Familial risk and clustering of nasopharyngeal carcinoma in Guangdong, China. Cancer 2004;101:363-9. [Crossref] [PubMed]
- Yu MC, Yuan JM. Epidemiology of nasopharyngeal carcinoma. Semin Cancer Biol 2002;12:421-9. [Crossref] [PubMed]
- Morrison JA, Gulley ML, Pathmanathan R, et al. Differential signaling pathways are activated in the Epstein-Barr virus-associated malignancies nasopharyngeal carcinoma and Hodgkin lymphoma. Cancer Res 2004;64:5251-60. [Crossref] [PubMed]
- Chou J, Lin YC, Kim J, et al. Nasopharyngeal carcinoma--review of the molecular mechanisms of tumorigenesis. Head Neck 2008;30:946-63. [Crossref] [PubMed]
- Hildesheim A, Apple RJ, Chen CJ, et al. Association of HLA class I and II alleles and extended haplotypes with nasopharyngeal carcinoma in Taiwan. J Natl Cancer Inst 2002;94:1780-9. [Crossref] [PubMed]
- Xiong W, Zeng ZY, Xia JH, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res 2004;64:1972-4. [Crossref] [PubMed]
- Zhang EP, Lian PG, Cai KL, et al. Radiation therapy of nasopharyngeal carcinoma: prognostic factors based on a 10-year follow-up of 1302 patients. Int J Radiat Oncol Biol Phys 1989;16:301-5. [Crossref] [PubMed]
- Vokes EE, Liebowitz DN, Weichselbaum RR. Nasopharyngeal carcinoma. Lancet 1997;350:1087-91. [Crossref] [PubMed]
- Hildesheim A, Anderson LM, Chen CJ, et al. CYP2E1 genetic polymorphisms and risk of nasopharyngeal carcinoma in Taiwan. J Natl Cancer Inst 1997;89:1207-12. [Crossref] [PubMed]
- Kongruttanachok N, Sukdikul S, Setavarin S, et al. Cytochrome P450 2E1 polymorphism and nasopharyngeal carcinoma development in Thailand: a correlative study. BMC Cancer 2001;1:4. [Crossref] [PubMed]
- Zeng ZY, Zhou YH, Zhang WL, et al. Gene expression profiling of nasopharyngeal carcinoma reveals the abnormally regulated Wnt signaling pathway. Hum Pathol 2007;38:120-33. [Crossref] [PubMed]
- Zeng Z, Zhou Y, Xiong W, et al. Analysis of gene expression identifies candidate molecular markers in nasopharyngeal carcinoma using microdissection and cDNA microarray. J Cancer Res Clin Oncol 2007;133:71-81. [Crossref] [PubMed]
- Fung LF, Lo AK, Yuen PW, et al. Differential gene expression in nasopharyngeal carcinoma cells. Life Sci 2000;67:923-36. [Crossref] [PubMed]
- Ang J, Lijovic M, Ashman LK, et al. CD151 protein expression predicts the clinical outcome of low-grade primary prostate cancer better than histologic grading: a new prognostic indicator? Cancer Epidemiol Biomarkers Prev 2004;13:1717-21. [PubMed]
- Tokuhara T, Hasegawa H, Hattori N, et al. Clinical significance of CD151 gene expression in non-small cell lung cancer. Clin Cancer Res 2001;7:4109-14. [PubMed]
- Wright MD, Moseley GW, van Spriel AB. Tetraspanin microdomains in immune cell signalling and malignant disease. Tissue Antigens 2004;64:533-42. [Crossref] [PubMed]
- Hemler ME. Targeting of tetraspanin proteins--potential benefits and strategies. Nat Rev Drug Discov 2008;7:747-58. [Crossref] [PubMed]
- Silvie O, Charrin S, Billard M, et al. Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites. J Cell Sci 2006;119:1992-2002. [Crossref] [PubMed]
- Gesierich S, Berezovskiy I, Ryschich E, et al. Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res 2006;66:7083-94. [Crossref] [PubMed]
- Takeda Y, Kazarov AR, Butterfield CE, et al. Deletion of tetraspanin Cd151 results in decreased pathologic angiogenesis in vivo and in vitro. Blood 2007;109:1524-32. [Crossref] [PubMed]
- Yang XH, Richardson AL, Torres-Arzayus MI, et al. CD151 accelerates breast cancer by regulating alpha 6 integrin function, signaling, and molecular organization. Cancer Res 2008;68:3204-13. [Crossref] [PubMed]
- Herlevsen M, Schmidt DS, Miyazaki K, et al. The association of the tetraspanin D6.1A with the alpha6beta4 integrin supports cell motility and liver metastasis formation. J Cell Sci 2003;116:4373-90. [Crossref] [PubMed]
- Zhou Z, Ran YL, Hu H, et al. TM4SF3 promotes esophageal carcinoma metastasis via upregulating ADAM12m expression. Clin Exp Metastasis 2008;25:537-48. [Crossref] [PubMed]
- Boucheix C, Soria C, Mirshahi M, et al. Characteristics of platelet aggregation induced by the monoclonal antibody ALB6 (acute lymphoblastic leukemia antigen p 24). Inhibition of aggregation by ALB6Fab. FEBS Lett 1983;161:289-95. [Crossref] [PubMed]
- Berthier-Vergnes O, El Kharbili M, de la Fouchardiere A, et al. Gene expression profiles of human melanoma cells with different invasive potential reveal TSPAN8 as a novel mediator of invasion. Br J Cancer 2011;104:155-65. [Crossref] [PubMed]
- Anami K, Oue N, Noguchi T, et al. TSPAN8, identified by Escherichia coli ampicillin secretion trap, is associated with cell growth and invasion in gastric cancer. Gastric Cancer 2016;19:370-80. [Crossref] [PubMed]
- Guo Q, Xia B, Zhang F, et al. Tetraspanin CO-029 inhibits colorectal cancer cell movement by deregulating cell-matrix and cell-cell adhesions. PLoS One 2012;7:e38464. [Crossref] [PubMed]
- Rana S, Claas C, Kretz CC, et al. Activation-induced internalization differs for the tetraspanins CD9 and Tspan8: Impact on tumor cell motility. Int J Biochem Cell Biol 2011;43:106-19. [Crossref] [PubMed]
- Qian CN, Berghuis B, Tsarfaty G, et al. Preparing the "soil": the primary tumor induces vasculature reorganization in the sentinel lymph node before the arrival of metastatic cancer cells. Cancer Res 2006;66:10365-76. [Crossref] [PubMed]
- Claas C, Seiter S, Claas A, et al. Association between the rat homologue of CO-029, a metastasis-associated tetraspanin molecule and consumption coagulopathy. J Cell Biol 1998;141:267-80. [Crossref] [PubMed]
- Kuhn S, Koch M, Nubel T, et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res 2007;5:553-67. [Crossref] [PubMed]
- Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci 2003;28:106-12. [Crossref] [PubMed]
- Seigneuret M, Delaguillaumie A, Lagaudriere-Gesbert C, et al. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J Biol Chem 2001;276:40055-64. [Crossref] [PubMed]
- Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci 2001;58:1189-205. [Crossref] [PubMed]
- Yue S, Mu W, Erb U, et al. The tetraspanins CD151 and Tspan8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget 2015;6:2366-84. [Crossref] [PubMed]
- Aggarwal BB, Shishodia S, Sandur SK, et al. Inflammation and cancer: how hot is the link? Biochem Pharmacol 2006;72:1605-21. [Crossref] [PubMed]
- Arlt A, Vorndamm J, Muerkoster S, et al. Autocrine production of interleukin 1beta confers constitutive nuclear factor kappaB activity and chemoresistance in pancreatic carcinoma cell lines. Cancer Res 2002;62:910-6. [PubMed]
- Saijo Y, Tanaka M, Miki M, et al. Proinflammatory cytokine IL-1 beta promotes tumor growth of Lewis lung carcinoma by induction of angiogenic factors: in vivo analysis of tumor-stromal interaction. J Immunol 2002;169:469-75. [Crossref] [PubMed]
- Al-toub M, Almusa A, Almajed M, et al. Pleiotropic effects of cancer cells' secreted factors on human stromal (mesenchymal) stem cells. Stem Cell Res Ther 2013;4:114. [Crossref] [PubMed]
- Feng X, Szulzewsky F, Yerevanian A, et al. Loss of CX3CR1 increases accumulation of inflammatory monocytes and promotes gliomagenesis. Oncotarget 2015;6:15077-94. [Crossref] [PubMed]
- Wang Z, Li P, Zhang Q, et al. Interleukin-1beta regulates the expression of glucocorticoid receptor isoforms in nasal polyps in vitro via p38 MAPK and JNK signal transduction pathways. J Inflamm (Lond) 2015;12:3. [Crossref] [PubMed]
- Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 2011;75:50-83. [Crossref] [PubMed]
- Cheng JQ, Godwin AK, Bellacosa A, et al. AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc Natl Acad Sci U S A 1992;89:9267-71. [Crossref] [PubMed]
- Yuan ZQ, Sun M, Feldman RI, et al. Frequent activation of AKT2 and induction of apoptosis by inhibition of phosphoinositide-3-OH kinase/Akt pathway in human ovarian cancer. Oncogene 2000;19:2324-30. [Crossref] [PubMed]
- Bellacosa A, de Feo D, Godwin AK, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer 1995;64:280-5. [Crossref] [PubMed]
- Zhang D, Brodt P. Type 1 insulin-like growth factor regulates MT1-MMP synthesis and tumor invasion via PI 3-kinase/Akt signaling. Oncogene 2003;22:974-82. [Crossref] [PubMed]


