Transcriptional dissection of differentially expressed long non-coding RNAs and messenger RNAs reveals the potential molecular mechanism after kidney transplantation
Introduction
Kidney transplantation is the most effective therapeutic option for end stage renal disease and provides a high-quality of life to these patients (1,2). In the past several decades, the short-term graft survival after operation has been significantly improved with the use of potent immunosuppressants (3,4). However, the long-term survival of kidney grafts remains to be advanced and is strongly threatened by cell-mediated rejection, antibody-mediated rejection (5,6) and other nonimmune factors, such as age, sex of donors and recipients, surgical operation, delayed graft function, postsurgical inflammation and oxidative stress, and drug side effects (7-10). Immunological rejection after transplantation is the issue that has received the most attention in this area, and several studies have highlighted immunosuppressants that prevent graft rejection and prolong graft survival (11-13). In contrast, little is known about the gene expression changes after kidney transplantation and after the start of immunosuppressants. The genetic mechanisms and pathways related to post operation and drug administration remain to be explored.
Long non-coding RNAs (lncRNAs) are a class of long non-protein-coding RNAs that modulate gene expression through a variety of mechanisms, which have not yet been clearly explored (14,15). In addition, the functions of most lncRNAs are still unknown (16). LncRNAs are as poorly conserved as the introns of coding genes and are less conserved than the 5'- or 3'-untranslated regions (UTRs) of messenger RNAs (mRNAs) (17). Given the relationship of lncRNAs and mRNAs, it is important to integrate these datasets to reveal the regulatory mechanisms among lncRNA-mRNA (18). To research the role of lncRNAs in genetic mechanisms related to kidney transplantation and to reveal the regulatory relationship among lncRNAs and mRNAs after transplantation and immunosuppressant administration, we analyzed the potential target protein-coding genes and lncRNAs and then performed an integrated analysis.
A previous study (19) conducted whole transcriptome sequencing in kidney transplantation and characterized changes in expression at multiple times posttransplantation. However, the work was limited in the discovery of differentially expressed mRNAs and the pathways involved in each time point after transplantation and pretransplantation. In this study, we reannotated the previous database to a more complete human genome, performed principal component analysis (PCA), compared the differentially expressed genes (DEGs) among all time points, researched the temporal variation of DEGs and constructed a lncRNA-mRNA network. All our results shed light on the gene variation profiles of posttransplantation and reveal the potential gene regulation mechanism after kidney transplant and immunosuppressant administration.
Methods
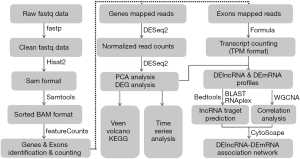
The flow chart of bioinformatics analyses is presented in Figure 1. The transcriptome data (GSE86884, NCBI GEO database) of peripheral blood mononuclear cells (PBMCs) isolated from whole blood samples with kidney transplantation recipients were used and were obtained by the RNA-seq technique on an Illumina HiSeq 2000 platform. A total of 32 kidney transplantation recipients were included in this study. None of them developed acute rejection during the 6-month study period. Peripheral blood samples were sequentially obtained pre-transplant (n=32), week 1 (n=31), months 3 (n=18) and 6 (n=15) post-transplant. Some samples were not collected because the patients were not seen for follow-up visits. Approximately 12 mL of blood per sample was collected into BD Vacutainer EDTA coated tubes for isolation of PBMCs. RNA was extracted using Qiagen QIAamp RNA Blood Mini kit (Germantown, MD) from PBMCs and evaluated for quantity and integrity. One ug quality of RNA with RNA integrity number greater than 6 was used to prepare polyA stranded RNAseq libraries.
Quality control and data processing
The acquisition of RNA-seq data included several steps: (I) obtaining raw reads: we downloaded raw fastq data from NCBI GEO database. Quality control and filtering of fastq data were performed using fastp with default settings (20). Tool fastp was a novel multifunctional, and ultra-fast FASTQ preprocessor with useful quality control and data-filtering features. (II) Read alignments: paired-end reads were then mapped to the human genome (hg38, UCSC) via the Hisat2 tool (version 2.1.0) (21). Tool Hisat2, a successor to both Hisat and Tophat2, exports alignments in SAM format. The authors have recommended that the Hisat/Tophat2 users switch to Hisat2. (III) Genes and exons quantification: before calculating quantification values, converting the SAM files were converted to BAM files and sorted by SAMtools (version 1.9) (22). The raw counts of genes and exons were determined using the featureCounts function from the Subread package (version 1.6.3) (23) where the gencode.v28.annotation.gtf file was used as a reference annotation guide. Transcripts per million reads (TPM) for each transcript were obtained from the raw counts of exons. At that time, we obtained quantification values of genes (read counts) or transcript (TPM).
Analysis of DEGs and time series analysis
Although some time series analysis tools are available, they are still in their early stages. We used the classical pairwise comparison approach with the R package DESeq2 (24). The overall gene expression profiles of the four time points (pretransplant and 1 week, 3 months, and 6 months posttransplant) were summarized through PCA. The DEGs were identified at 1 week vs. pretransplant (as 2 vs. 1 group), 3 months vs. 1 week (as 3 vs. 2 group), 6 vs. 3 months (as 4 vs. 3 group) and 6 months vs. pretransplant (as 4 vs. 1 group), with the statistical significance threshold at a false discovery rate (FDR) <0.05 and a fold change ≥2 in the analysis. These genes were restricted to the top 50% of the mean expression level to remove low levels of genes. DEGs were displayed by Veen and volcano plots and further investigated for function and pathway enrichment using KEGG analysis.
LncRNA target prediction and construction of a lncRNA-mRNA regulatory network
Based on the expression level of log2(TPM+1), we first calculated the correlation coefficient and P between arbitrary differentially expressed lncRNA (DElncRNA) and arbitrary differentially expressed mRNA (DEmRNA) in the 2 vs. 1 group, respectively. Next, lncRNA target predictions were superimposed onto the lncRNA-mRNA correlation network to suggest whether the lncRNAs directly regulated the expression of the target mRNAs. Similar to Han’s study, DElncRNAs were chosen for target prediction to identify the cis- or trans-regulatory effects (25). Tool BEDtools was used to screen out cis target genes of each DElncRNA, i.e., DEmRNAs transcribed within a range of 10kb upstream or downstream of each DElncRNA. Tools BLAST and RNAplex were utilized to find out trans target genes of each DElncRNA based on sequence complementarity and RNA duplex energy prediction. The BLAST parameters were set as “-evalue 1e-5 -perc_identity 90”, and the RNAplex parameters were set as “-e -30”. The requirements for DElncRNA paired with DEmRNA were absolute correlation coefficient >0.5 and P<0.05 at any one of two time points (group 1 and 2). Finally, DElncRNA-DEmRNA regulatory network was constructed and visualized by CytoScape (version 3.4.0).
Results
DEGs during pre- and posttransplant
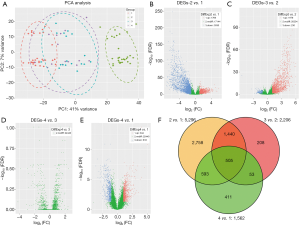
The results of PCA are shown in Figure 2A. The pretransplant, three-month and 6-month groups had a high overlap, whereas the one-week posttransplant group was different from the others based on the gene expression profiles. The volcano plot depicting global expression patterns indicated DEGs among the four groups (Figure 2B,C,D,E). After filtering out low count genes by DESeq2, Whether the remaining 24,440 genes were differentially expressed was our focus. Figure 2B shows that 1,766 genes were upregulated at 1 week after kidney transplantation compared to pretransplant (time point 2 vs. 1), whereas 3,530 genes were downregulated. In order to determine how 5,296 above-mentioned DEGs changed or whether there was a significant difference in the expression of other genes for the next 6 months, DEG analysis was performed in the 3 vs. 2 group, the 4 vs. 3 group and the 4 vs. 1 group. There were 2,206 DEGs in the 3 vs. 2 group, which is indicated in Figure 2C. Interestingly, there were no DEGs in the 4 vs. 3 group (Figure 2D). This reflected that the gene profile at 6 and 3 months after operation was roughly the same. However, a total of 1,562 genes were expressed differentially 6 months after surgery as opposed to that pretransplant (Figure 2E). The results suggested that the number of DEGs declined over time. Comparing the significant DEGs among the 2 vs. 1, 3 vs. 2 and 4 vs. 1 group, there were 505 common DEGs. A Venn diagram of gene expression analysis represented the overlap of the three groups (Figure 2F). These results suggested that the number of DEGs declined and gene expression tended to be stable over time.
Time series trend of DEGs after kidney transplantation
In order to identify the dynamic change of these genes at four different time points, we extracted 5,557 DEGs in the 2 vs. 1 or 3 vs. 2 groups, which were differentially expressed at least once, to construct a time series trend analysis. These genes were divided into eight clusters according to upregulation, downregulation and nondifference among the 2 vs. 1 and 3 vs. 2 groups without double nondifference. Then, we defined the trend as upregulation or downregulation based on the change folds in the 4 vs. 3 group, although the FDR was nonsignificant. In general, we separated these DEGs into 16 clusters, and the temporal variation expression figures were established on the average expression of DEGs in each cluster in the samples at each time point.
As shown in Figure 3, the expression levels of gene sets were increased at 1 week posttransplant and are presented in clusters 1–6 (Figure 3A,B,C,D,E,F). Some of the genes in clusters 1–2 continued to increase at 3 months compared to 1 week, suggesting the existence of a long-term effect. For the early recovery genes in clusters 3–4, the expression dropped to the level of pretransplant at 3 months. There was no significant change at 3 months compared to 1 week in clusters 5–6, but most of these genes appeared to decrease within a period of 1 week to 6 months after transplantation. Clusters 7–12 contained genes that were downregulated at 1 week compared to pretransplant (Figure 3G,H,I,J,K,L). The genes in clusters 7–8 appeared early in recovery, and a long-term affected trend was observed in clusters 9–10. The expression of the genes that changed at the later period instead of 1 week posttransplant were in clusters 13–16, which suggested a chronic potential effect of immunosuppressants (Figure 3M,N,O,P). A brief table is shown in Figure 3Q. We also performed DEG analysis on the 4 vs. 1 group. By overlapping the results with the above 5,557 genes, 1,151 genes were differentially expressed compared to that pretransplant (Figure 2F). These results suggested the likely effects of surgery and drug administration.

KEGG pathway analysis
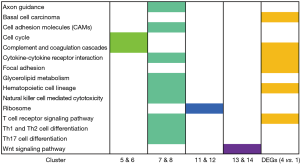
We took every two clusters for KEGG pathway analysis (Figure S1). The results showed that clusters 7 & 8 involved immune mechanism pathways, such as the T cell receptor signaling pathway, Th1 and Th2 cell differentiation, and cytokine-cytokine receptor interaction. Complement and coagulation cascades and the cell cycle pathway had a significant effect in clusters 5 & 6. The ribosome pathway played a major role in clusters 11 & 12, whereas the Wnt signaling pathway was the top pathway related to clusters 13 & 14. The pathway analysis suggested that the attenuation of the immune reaction, reduced expression of ribosome-related proteins and the change in the PBMC cell cycle occurred after the use of immunosuppressants.
Construction of lncRNA-mRNA association network
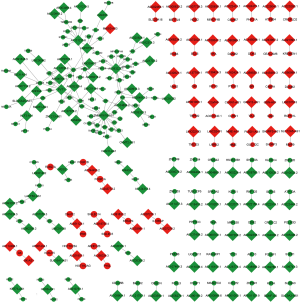
With the above methods, we constructed an association network including the lncRNAs and target genes that were differentially expressed in the 2 vs. 1 or 3 vs. 2 groups (Figure 4). There were 1,063 DElncRNAs and 3,563 DEmRNAs revealed by the correlation analysis, and the results showed that 40,432 lncRNA-mRNA pairs simultaneously met an absolute correlation coefficient >0.5 and P<0.05 at two time points. In addition, we obtained 466 lncRNA-mRNA pairs with cis-regulation and 5,692 pairs with trans-regulation. Combined with the restriction condition of the correlation analysis results, the network had 308 nodes and 235 connections between 138 lncRNAs and 170 mRNAs. This network indicated that lncRNA TMEM161B-AS1 targeted eighteen mRNAs (such as ANGEL1, ARSK, CARF, CATSPER2, CD96, DIEXF, ELP2, HLCS) in the 2 vs. 1 group. ZBTB25 was a target gene of seven lncRNAs (AC007114.2, AL122035.2, AL591848.3, IQCH-AS1, KLF3-AS1, PVT1, SLC25A25-AS1). Several single lncRNAs targeted single mRNAs. A portion of the targets mentioned here have been reported to be linked to cancer and transplantation. These results suggested a potential regulatory mechanism of lncRNAs to mRNAs after kidney transplantation.
Conclusions
The benefits of transplantation for end-stage renal disease patients have attracted much attention from academia, and great progress has been made in basic and clinical research on kidney transplantation. Although routine use of immunosuppressive drugs after operation have reduced the occurrence of acute rejection effectively, but chronic allograft injury limited the improvement in long-term allograft survival. Whether in case of acute rejection or chronic allograft injury, to study the genetic changes before pathological changes and functional changes was crucial. Most previous studies have focused on DEGs at the time of an acute rejection or chronic allograft injury event, but the small number of samples and widely varied baseline data made lower the credibility of its research results.
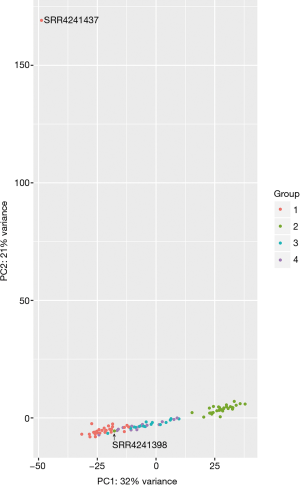
Researches on PBMCs is essential in the process of graft rejection after kidney transplantation beginning with lymphocytes infiltration. The RNA sequencing data were obtained from previous work, in which 32 patients receiving living donor kidney allografts were involved. The research has investigated changes in gene expression after kidney transplantation in patients without acute rejection or chronic allograft injury. This was a good reference for the screening of biomarkers predicting an acute rejection or chronic allograft injury event. However, there were several aspects that made our research different from or more rigorous than the research of the original data provider. Firstly, we mapped raw fastq data to the latest human genome (GRCh38/hg38) that covered a lot of information about non-coding genes. We picked out 1,063 DElncRNAs from 5,557 DEGs either in group 2 vs.1 or group 3 vs. 2 for further studies. Secondly, we eliminated two samples that were far away from the samples-set by PCA analysis (SRR4241437 and SRR4241398, Figure S2). It made the results more reliable and credible. The results explained the benefits of kidney transplantation to patients, particularly at the later stage, compared to other therapies (26,27).
Thirdly, all of our analysis was conducted by the current mainstream of data-analysis softwares and methods, such as, Hisat2 instead of Hisat/Tophat2 in read alignments, TPM instead of FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) in gene quantification, and three programs (BEDtools, BLAST and RNAplex) in DElncRNA-target prediction. Fourthly, the comparative objects of DEG analysis were different. We did DEG analysis in the 2 vs. 1 group, the 3 vs. 2 groups, the 4 vs. 3 groups and the 4 vs. 1 group, but original authors focused on DEGs after transplant compared to baseline at pretransplant. They concluded that gene expression signatures at month 3 were similar to week 1. However, our study suggested gene expression signatures at week 1 posttransplant were the most different from that at other three time points, based on the result of PCA and time series analysis. They ignored that the dynamic change of DEGs. In our opinion, to observe that the dynamic change of each gene at different time points was more important than to compare the difference among different time points. For this reason, we constructed time series trend analysis by semi-artificial comparison instead of an immature but simple tool. Both our study and previous study clarified that the numbers of DEGs after transplant compared to baseline gradually decreased over time. Fifthly, to advance the study of gene mechanism especially in the reaction of lncRNAs to mRNAs before and after transplantation, we superimposed the result of DElncRNA-target prediction to that of correlation analysis for construction of the gene regulatory network. This was a unique analysis in our research, which did not exist in the previous study. Correlation analysis stated pairwise co-expressed genes with direct or indirect relationship, and DElncRNA-target prediction method identified a batch of directly acting potential target genes of one DElncRNA on basis of position and sequence structure. We eventually built several hundreds of DElncRNA-DEmRNA pairs that have yet to be verified.
In the last aspect, we paid attention to cell-type specific gene expression profiles. Most studies about whole genome expression still relied on the analysis of the total PBMCs (28-30). There were several cell subsets in PBMCs, so the information that PBMCs provided was more complete but heterogeneous: the differential expression might be due to the change of gene expression, the change of cell type proportions or both. According to the identification of specific gene expression signatures for B-cells (427 genes) and T-cells (222 genes), we explored the dynamic changes of these specific genes for reflecting the activity of humoral immunity and cellular immunity (31).
In our study, the number of T-cells specific DEGs was ninety; 75.6% (n=68) and 13.3% (n=12) were classified into cluster 7 & 8 and cluster 11 & 12 respectively. The expression level of those genes decreased at 1 week following transplant but increased towards pretransplant levels over time in the blood. Those genes encompassed T-cell receptor (e.g., CD3D, CD3E, CD3G and CD247), T-cell costimulatory molecule (e.g., CD28), signal transduction-related genes (e.g., CD96, FYN, LAT, MAL, TRAT1, TRGC1 and ZAP70), cell adhesion molecules (e.g., CD2, CD5, CD6) and genes encoding T-cell transcriptional regulators (e.g., GATA3, LEF1, STAT4 and TCF7). General down-regulation of above-mentioned several kinds of genes was probably because of depletion of T cells in the early stage after kidney transplantation. Only 7.8% (7) of T-cells specific DEGs were classified into cluster 6, including ACTN1, ARID3A, BEX3, OLAH, PKM, S100A8 and UPP1. ACTN1 was known to be involved in cytoskeletal remodeling and calcium mobilization, a fundamental process for T-cell activation, but it was recently described as a differentiation/maturation marker of CD8+ T-cell (32). Up-regulated PKM was related to enhanced Th1 and Th17 cell differentiation (33). The activation marker S100A8 was strongly increased in CD8+ T cells. In addition, the dynamic change of immune checkpoint molecules inhibiting T-cells was also assessed. Five checkpoint molecules (CD124/IL7R, CD152/CTLA4, CD160, CD223/LAG3 and TIGIT) were all classified into cluster 7. Other four checkpoint molecules (CD137, CD279/PD1, CD366/TIM3 and VISTA) were not included in all 5,557 DEGs. It showed that the immune effect of T cells was gradually suppressed by taking immunosuppressive drugs for a long time. On the other hand, the number of B-cells specific DEGs was forty-four. Six genes (CR1, IGHD, IGHG1, IGHG2, IGHG3 and IL4R) were involved in B cell mediated immunity. CR1 and IL4R were in cluster 6, and four immunoglobulin heavy genes were in cluster 11. The function of the rest of specific genes was not clear. Most of B-cell co-receptor molecules, B-cell surface markers, signal transduction molecules, and transcriptional regulators were non-different. Considering the differential subtypes in PBMCs, which lymphocytes are responsible for the altered transcripts and the relative frequency needs to be explored in the future.
To explore the dynamic changes of those DEGs, we performed a time-series trend analysis and identified 16 gene clusters. We have not retrieved other published research related to the temporal variation in gene expression in patients without rejection. The expression of genes in clusters 3, 4, 7, and 8 were changed at 1 week versus pretransplant and meaningfully returned to the trend of preoperation in 3 months, which was probably explained by the early effect of drugs to avoid acute rejection. It is interesting to research these genes, which significantly changed at 1 week but then recovered at 3 months by enlarging the effects of anti-acute rejection with drugs. Genes such as MMP9, CD177, IL-34, and LAMC3 were in clusters 5, 6, 11, and 12 and appeared to recover later at three and 6 months. Turunen et al. (34) reported that MMP9 is associated with cold ischemia time and the emergence of delayed graft function related to kidney transplantation. IL-34 was reviewed by Baghdadi et al. (35), and growing evidence has indicated a correlation between IL-34 and disease severity, chronicity and progression. Based on these findings, we hypothesized that the later-recovered genes may act as targets to attenuate chronic rejection and improve the life of grafts. The expression of genes in clusters 1–2 and 9–10 were upregulated or downregulated chronically post-transplantation, which suggested long-term damage by surgery or immunosuppressants. From the temporal variation, we clearly understand the influence of immunosuppressants, which resulted in the changes of genes in PBMCs. The DEGs in each cluster probably play different roles after kidney transplantation depending on the temporal variation, which remains to be explored.
We subsequently performed KEGG analysis on every similar cluster, and we found that clusters 7 & 8 focused on immune pathways, such as the T cell receptor signaling pathway, Th1 and Th2 cell differentiation, cytokine-cytokine receptor interaction and Th17 cell differentiation. The results suggested that the immune system was acutely inhibited after transplantation with the use of immunosuppressants and partially recovered after 3 months, establishing a new equilibrium in PBMCs. It supported the efforts and achievements of research in immunosuppressants; however, the posttransplantation immune reactions are still worth mentioning. The Wnt signaling pathway was the most enriched pathway in clusters 13 & 14, which was reported in T lymphocyte responses. Recent evidence indicated that the Wnt pathway serves as a master regulator of T cell immune responses by governing the balance between T helper 17/regulatory T cells and regulating the formation of effector and memory cytotoxic CD8 T cell responses, which influences the outcome of immune responses in transplantation (36). In general, the pathways from the clusters in this study revealed that the immune system was attenuated, the expression of ribosome-related proteins was downregulated and the cell cycle of PBMCs was changed by immunosuppressants after transplantation. Increasing evidence has confirmed that lncRNAs are one of the most important factors controlling gene expression (37). To explore the molecular mechanism after kidney transplantation, we combined DEmRNAs with target gene prediction of DElncRNAs. Finally, we obtained 235 credible lncRNA-mRNA pairs with cis- or trans-regulatory effects. This network indicated that TMEM161B-AS1 targeted the highest number of mRNAs with a positive regulatory relationship and that ZBTB25 was targeted by the largest number of lncRNAs. By regulating these lncRNAs, the expression of corresponding mRNA would be changed and play differential functions in the processes after transplantation. The lncRNA-mRNA network revealed a potential regulatory mechanism of lncRNAs targeting mRNAs after kidney transplantation.
In conclusion, our study reannotated the previous database to a more complete human genome, obtained more intricate DEGs at each time point after kidney transplantation, researched the temporal variation of those DEGs and constructed a lncRNA-mRNA network. This work exhibited a transcriptional profile based on the time course and revealed several hundred significantly altered lncRNA-mRNA axes contributing to molecular regulation after kidney transplantation, which helps us to understand the molecular mechanism after kidney transplantation and the effects of immunosuppressive drugs.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant numbers 81870512, 81570676, 81871980, 81902455), the “333 High Level Talents Project” in Jiangsu Province (grant number BRA2017532), the Standardized Diagnosis and Treatment Research Program of Key Diseases in Jiangsu Province, China (grant number BE2016791), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant number KYCX17_1253), the Clinical Advanced Technology Program of Jiangsu Science and Technology Agency (BE2016788), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, JX10231801).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research was based on the existing database, which was approved by the institutional review board of the University of Minnesota.
References
- Rana A, Gruessner A, Agopian VG, et al. Survival benefit of solid-organ transplant in the United States. JAMA Surg 2015;150:252-9. [Crossref] [PubMed]
- Garcia GG, Harden P, Chapman J, et al. The global role of kidney transplantation. Nephrol Dial Transplant 2013;28:e1-5. [Crossref] [PubMed]
- Nickel P, Bestard O, Volk HD, et al. Diagnostic value of T-cell monitoring assays in kidney transplantation. Curr Opin Organ Transplant 2009;14:426-31. [Crossref] [PubMed]
- Hariharan S, Stablein DE. Improvements in long-term renal transplant graft survival. American Journal of Transplantation 2005;5:630-1. [Crossref] [PubMed]
- Li L, Khush K, Hsieh SC, et al. Identification of common blood gene signatures for the diagnosis of renal and cardiac acute allograft rejection. PLoS One 2013;8:e82153. [Crossref] [PubMed]
- Issa F, Schiopu A, Wood KJ. Role of T cells in graft rejection and transplantation tolerance. Expert Rev Clin Immunol 2010;6:155-69. [Crossref] [PubMed]
- Głyda M, Włodarczyk Z, Czapiewski W. Results of renal transplantation from expanded criteria deceased donors - a single-center experience. Ann Transplant 2012;17:35-42. [Crossref] [PubMed]
- Tetaz R, Trocme C, Roustit M, et al. Predictive diagnostic of chronic allograft dysfunction using urinary proteomics analysis. Ann Transplant 2012;17:52-60. [Crossref] [PubMed]
- Pita-Fernández S, Valdés-Cañedo F, Seoane-Pillado T, et al. Influence of early graft function after renal transplantation and its impact on long-term graft and patient survival. Transplant Proc 2010;42:2856-8. [Crossref] [PubMed]
- Salamzadeh J, Sahraee Z, Nafar M, et al. Delayed graft function (DGF) after living donor kidney transplantation: a study of possible explanatory factors. Ann Transplant 2012;17:69-76. [Crossref] [PubMed]
- Bouatou Y, Viglietti D, Pievani D, et al. Response to Treatment and Long-Term Outcomes in Kidney Transplant Recipients with Acute T Cell-Mediated Rejection. Am J Transplant 2019;19:1972-88. [PubMed]
- Hoffman W, Mehta R, Jorgensen DR, et al. The Impact of Early Clinical and Subclinical T Cell Mediated Rejection after Kidney Transplantation. Transplantation 2019;103:1457-67. [Crossref] [PubMed]
- McAdams-DeMarco MA, Law A, Tan J, et al. Frailty, mycophenolate reduction, and graft loss in kidney transplant recipients. Transplantation 2015;99:805-10. [Crossref] [PubMed]
- Whitehead J, Pandey GK, Kanduri C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta 2009;1790:936-47.
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev 2005;19:1635-55. [Crossref] [PubMed]
- Li Y, Egranov SD, Yang L, et al. Molecular Mechanisms of Long Noncoding RNAs-mediated Cancer Metastasis. Genes Chromosomes Cancer 2019;58:200-7. [Crossref] [PubMed]
- Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet 2006;22:1-5. [Crossref] [PubMed]
- Li Z, Xiong C, Mo S, et al. Comprehensive Transcriptome Analyses of the Fructose-Fed Syrian Golden Hamster Liver Provides Novel Insights into Lipid Metabolism. PLoS One 2016;11:e0162402. [Crossref] [PubMed]
- Dorr C, Wu B, Guan W, et al. Differentially expressed gene transcripts using RNA sequencing from the blood of immunosuppressed kidney allograft recipients. PLoS One 2015;10:e0125045. [Crossref] [PubMed]
- Chen S, Zhou Y, Chen Y, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34:i884-90. [Crossref] [PubMed]
- Conesa A, Madrigal P, Tarazona S, et al. A survey of best practices for RNA-seq data analysis. Genome Biol 2016;17:13. [Crossref] [PubMed]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078-9. [Crossref] [PubMed]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923-30. [Crossref] [PubMed]
- Spies D, Renz PF, Beyer TA, et al. Comparative analysis of differential gene expression tools for RNA sequencing time course data. Brief Bioinform 2019;20:288-98. [Crossref] [PubMed]
- Han L, Zhang K, Shi Z, et al. LncRNA pro fi le of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol 2012;40:2004-12. [PubMed]
- Chertow GM, Levin NW, Beck GJ, et al. Long-Term Effects of Frequent In-Center Hemodialysis. J Am Soc Nephrol 2016;27:1830-6. [Crossref] [PubMed]
- Campbell D, Mudge DW, Craig JC, et al. Antimicrobial agents for preventing peritonitis in peritoneal dialysis patients. Cochrane Database Syst Rev 2017;4:CD004679. [PubMed]
- Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant 2004;4:1475-89. [Crossref] [PubMed]
- Li L, Khatri P, Sigdel TK, et al. A peripheral blood diagnostic test for acute rejection in renal transplantation. Am J Transplant 2012;12:2710-8. [Crossref] [PubMed]
- Kurian SM, Williams AN, Gelbart T, et al. Molecular classifiers for acute kidney transplant rejection in peripheral blood by whole genome gene expression profiling. Am J Transplant 2014;14:1164-72. [Crossref] [PubMed]
- Palmer C, Diehn M, Alizadeh AA, et al. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics 2006;7:115. [Crossref] [PubMed]
- Costa M, Cruz E, Oliveira S, et al. Lymphocyte gene expression signatures from patients and mouse models of hereditary hemochromatosis reveal a function of HFE as a negative regulator of CD8+ T-lymphocyte activation and differentiation in vivo. PLoS One 2015;10:e0124246. [Crossref] [PubMed]
- Baĭzhomartov MS, Prozorovskiĭ SV, Irzhanov SD, et al. Hypersensitivity of the delayed type in immunization and infection with Mycoplasma pneumoniae. Zh Mikrobiol Epidemiol Immunobiol 1975.75-8. [PubMed]
- Turunen AJ, Lindgren L, Salmela KT, et al. Matrix Metalloproteinase-9 and Graft Preservation Injury in Clinical Renal Transplantation. Transplant Proc 2015;47:2831-5. [Crossref] [PubMed]
- Baghdadi M, Endo H, Tanaka Y, et al. Interleukin 34, from pathogenesis to clinical applications. Cytokine 2017;99:139-47. [Crossref] [PubMed]
- Staal FJ, Arens R. Wnt Signaling as Master Regulator of T-Lymphocyte Responses: Implications for Transplant Therapy. Transplantation 2016;100:2584-92. [Crossref] [PubMed]
- Khachane AN, Harrison PM. Mining mammalian transcript data for functional long non-coding RNAs. PLoS One 2010;5:e10316. [Crossref] [PubMed]