
Long non-coding RNAs in diseases related to inflammation and immunity
Introduction
The immune system consists different cell types that mediate the response to infection and maintain a dynamic balance in the body. While maintaining the dynamic balance, the external environment of each cell affects its appearance and transcription spectrum for immunity. Many immune system diseases have unknown pathogenic factors, and no effective treatment has yet been found. At present, non-coding RNAs have been a research hotspot and difficulty. New evidence suggests that long non-coding RNAs (lncRNAs) play a key role in epigenetic and transcriptional regulation in mammals, which regulate diverse biological functions including cell differentiation, cell lineage choice, organogenesis and tissue homeostasis. In innate and adaptive immunity, lncRNAs have been key regulators of gene expression in immune cells, however, the mechanism of how lncRNAs regulate immune function remains unclear. Here we review lncRNAs in the control of cellular function and homeostasis in immune response, the different molecular mechanisms in regulating transcription processes, and the involvement in related diseases to provide new ideas for future research and treatment of immune-related diseases.
Structure and function of lncRNAs
LncRNAs, non-protein-encoding transcripts, were discovered by whole transcriptome sequencing, are greater than 200 bp in length, have a polyadenylate 5' cap structure, and are polyadenylated like mRNAs. The main difference between lncRNA and mRNA is that lncRNA contains transcriptional termination codons, therefore lncRNA does not have any protein coding potential (1). Humans contain approximately the same number of protein-coding genes (~20,000) as well as other vertebrates, such as nematodes and roundworms (2). However, the length of genes is different among various species. In fact, a part of the human genome is used to synthesize lncRNA during evolution. The lower the evolutionary level of species, such as zebrafish, nematodes, fruit flies and yeasts, the lower the number of lncRNAs and ratio of lncRNAs to protein-coding genes (3,4). LncRNAs play an important role in many developmental processes and exhibit cell type specific in expression pattern and function. It is speculated that the complexity of species needs more lncRNA genes (5-7).
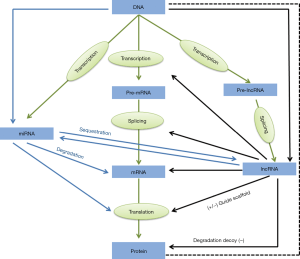
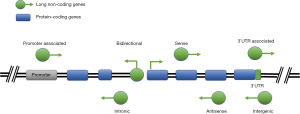
LncRNAs are usually classified and named according to their position in genomes relative to protein-coding genes and enhancer regulatory elements, including long intergenic non-coding RNA (lincRNA), intronic transcript, antisense (AS) lncRNA and enhancer RNA (eRNA) and so on (Figure 1). The lncRNAs between two protein-coding genes that do not overlap with the protein-coding genes are usually abbreviated as lincRNAs, which refer to the long or large interval ncRNAs or inserted ncRNAs; lncRNAs can also be located in an intron of the protein-coding gene without overlapping with the exon, known as “Intronic”; some lncRNA genes overlap with the exons and introns of the protein-coding genes, but these new exons are not located in the protein-coding genes, and are usually transcribed from AS DNA strands, known as AS lncRNAs. However, the number of lncRNAs studied in detail is relatively small. The common mechanisms for lncRNAs involve DNA, protein or RNA (8). LncRNAs act as following: (I) by transcription in the upstream promoter region of the protein-coding genes to interfere the expression of adjacent protein-coding genes; (II) by inhibiting RNA polymerase, or mediating remodeling of chromatin and modification of histone to affect gene expression; (III) by complementing the double strand with the transcripts of protein-coding genes to interfere the splicing of mRNA; (IV) by complementing the double strand with the transcripts of protein-coding genes to product endogenous small interference RNA by dicer enzyme to regulate gene expression; (V) by binding to the specific protein to regulate the activity; (VI) by forming a nucleic acid protein complex with a protein as a structural component; (VII) by binding to the specific protein to alter cytoplasmic localization of the protein; (VIII) as precursor molecules of small RNAs [e.g., microRNA, PIWI-interacting small RNA (piRNA), and MALAT1-associated small cytoplasmic RNA (mascRNA)]. In view of the powerful function of human lncRNAs, their mechanisms still need to be continuously explored (Figure 2).
LncRNA and immunity
Many studies revealed that lncRNAs showed a viability and cell specific in proliferation, differentiation and activation of immune cells, including monocytes, macrophages, dendritic cells (DC), neutrophils, T cells and B cell (8). For example, non-coding transcripts of nuclear factor of activated T-cells (NFAT) (NRON) can inhibit the transcriptional activity of human NFAT, and bring the phosphorylated NFAT into a cytoplasmic RNA-protein complex as a “scaffolding” tool (9,10); lincR-Ccr2-5'AS, as a Th2-specific lncRNA, can regulate the expression of immune-related genes in Th2 cells and move mouse Th2 cells to the lungs (11); Tmevpg1 can be co-expressed with Ifng, and can stimulate epigenetic remodeling with Ifng-specific enhancers by T-bet to achieve specific expression of Th1 family member, Ifng (12,13); linc-MAF-4 is a CD4+ Th1 cell-specific lncRNA, which promotes the differentiation of CD4+ T cells into Th1 cells, and regulates the transcription of MAF as a scaffold (14); lnc-DC regulates the differentiation of human monocytes into DC, directly interacts with signal transducer and activator of transcription 3 (STAT3) in the cytoplasm, and prevents the dephosphorylation of STAT3 from SHP1 (15); HOTAIRM1 is specifically expressed in bone marrow cells, and induces promyelocytic NB4 cells into leukemia cells by retinoic acid, and is up regulated during differentiation from normal human hematopoietic stem cells to granulocytes (16). Transcriptome analysis in CD8+ and CD4+ T cells identified a large number of lncRNAs and showed stage- or tissue-specific expression (17,18). However, the development and function of lncRNAs in B cells are poorly understood.
The role of lncRNA in innate immunity
Innate immunity including macrophages, DC, neutrophils, and acidophilic/basic granulocytes, is the first line against microbial pathogens. Recent studies have shown that lncRNAs such as lincRNA-Cox2, Lethe, PACER and THRIL, members of the lncRNA family play an important role in the control of gene expression in immune cells (Figure 3).
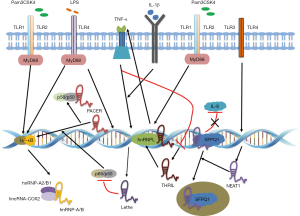
By RNA sequencing of bone marrow-derived macrophages (BMDM), 72 lncRNAs were found to be significantly up-regulated in macrophages exposed to Pam3CSK4, which was related to synthetic bacterial lipoproteins (19). One of the most classical lncRNAs (lincRNA-Cox2) has been confirmed to be a regulator of the transcription of macrophages Toll-like receptor (TLR) (20). It has been reported that the lincRNA-Cox2 gene was classified as a lincRNA, which was able to activate or inhibit the expression of different types of immune genes. In resting macrophages, lincRNA-Cox2 inhibits the expression of approximately 700 genes, including chemokines (Ccl5 and Cx3cl1) and interferon-stimulated genes (ISG) (Irf7, Isg15, Ifi204, and Oas2), while it can also activate various genes (IL-6, Tlr1 and IL-23a) via the TLR2 pathway. LincRNA-Cox2 interacts with RNA binding protein (RBP) heterogeneous nuclear ribonucleoprotein (hnRNP)-A2/B1 and hnRNP-A/B to regulate its inhibitory function. Interestingly enough, these two hnRNPs are not involved in the lncRNA-mediated activation, suggesting that there are other unknown proteins that interact with lincRNA-Cox2. The specific role of lincRNA-Cox2 on TLR2-induced IL-6 and other gene expression is currently unknown (21).
Lethe, a functional pseudogene (Rps15a-PS4) lncRNA, is also highly induced in mouse embryonic fibroblasts (MEFs) treated by nuclear factor kappa B (NF-κB)-activated inflammation that triggers TNFα and IL-1β expression. Lethe also responds to the adrenal cortex hormone and dexamethasone. Lethe also contacts with p65 (RelA) to attenuate NF-κB-dependent inflammatory responses, which prevent RelA from binding to the promoter region of target genes, including IL-6, IL-8, and superoxide dismutase 2 (SOD2). Interestingly, the expression of Lethe in the spleens of older rats was significantly lower (20- to 50-fold) than that in younger mice, suggesting that the decrease in Lethe expression is associated with increased NF-κB signaling related to aging (22). Therefore, Lethe can act as a feedback regulator of the NF-κB signaling pathway to control the inflammatory response. More importantly, it provides an evidence for the presence of more functional pseudogenes lncRNA in the mammalian genome. In addition, lncRNA P50-associated COX-2 extragenic RNA (PACER) may also be a regulator of the NF-κB signaling pathway. In the lipopolysaccharide (LPS)-stimulated human macrophages, PACER was up regulated and selectively regulated the expression of the adjacent gene PTGS2 (COX-2) (23).
THRIL is another immune-related lncRNA that primarily controls the expression of TNFα in the human monocyte cell line THP-1. In Pam3CSK4-treated THP-1 cells, THRIL was identified as one of the differentially expressed 159 lincRNAs. Nine of the lincRNAs (including THRIL) knocked down by lentiviral short hairpin RNA (shRNA) caused the damage to TNFα and/or IL-6 products, suggesting that some TLR2-induced lincRNAs were involved in the regulation of human monocytes Inflammation. In Pam3CSK4-treated THP-1 cells, RNA-seq showed 319 differentially expressed genes after shRNA silenced THRIL, indicating that THRIL broadly regulated immune-related genes. THRIL interacts with hnRNPL to regulate TNFα expression, and both THRIL and hnRNPL sites in the TNFα promoter region. Since TNFα inhibited the expression of THRIL, THRIL has been a negative feedback regulator of TNFα expression in human monocytes (24).
Nuclear enriched abundant transcript 1 (NEAT1), as a key lncRNA in the control of heterochromatin structure formation, localizes SFPQ (A NEAT1 binding protein) into the nucleus to mediate IL-8 transcription after human cell lines are infected with virus (HSV-1 and influenza A virus) or activated by TLR3 (25). In addition, NEAT1 also transfers HIV-1 mRNA from the nucleus to the cytoplasm. Together, these studies illustrate the important role of lncRNAs in regulating gene expression in innate immunity (26).
Although many literatures described the regulatory role of lncRNAs in inflammatory gene expression, little has been known about the development and homeostatic regulation of innate immune cells. HOTAIRM1 is one of them, highly expressed in human granulocytes, and induced by retinoic acid signaling and PU.1 during myeloid differentiation (16,27). In vitro studies showed that knockdown of HOTAIRM1 led to decreased expression of CD11b and CD18 and impaired granulocyte differentiation (16). However, the role of HOTAIRM1 on the differentiation of human granulocytes in vivo remains unclear. Recent studies have also found that lncRNA Morrbid regulated the homeostasis of eosinophils, neutrophils and Ly6Chi monocytes by modulating the proapoptotic factor BCL2L11 (also known as Bim) (28). In myeloid cells, proinflammatory cytokines (such as IL-3, IL-5, GM-CSF, etc.) induce the expression of Morrbid, which can accumulate polycomb repressive complexes 2 (PRC2) on the BCL2L11 promoter to inhibit BCL2L11 transcription and promote the survival of the cells. In the absence of Morrbid, cell apoptosis is increased. Thus, there is a new and critical approach to precisely regulate the lifespan of these inflammatory cells. In fact, high expression of Morrbid is present in eosinophils in patients with hypereosinophilic syndrome (HES), which is characterized by the altered lifespan of eosinophils (28). Taken together, these data suggested that the Morrbid-BCL2L11 axis might be an important factor in the regulation of lifespan of myeloid cells in HES, inflammation and cancer.
The role of lncRNA in adaptive immunity
Adaptive immune means that the body produces an effective specific antigen-antibody reaction and forms long-term immune memory, while avoiding autoimmune and chronic inflammatory reactions, including T cells and B cells. Some evidence demonstrated that lymphocytes expressed a large number of lncRNAs and played a key role on development, differentiation and activation of cells. Two important lncRNAs expressed in T cells are the NTT, non-coding transcript in CD4+ T cells, and NRON, one of the earliest lncRNA genes identified in immune cells (Figure 4).
NTT is mainly expressed in activated human CD4+ T cells, but its function remains unknown. The potential relationship between NTT and interferon-gamma receptor (IFN-γR) also needs to be further validated, because they act on the same locus (6q23-q24) and show the analogous expression during T cell activation (9,29).
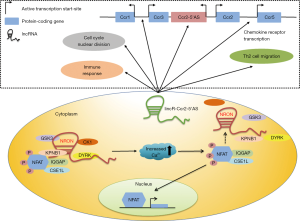
The lncRNA NRON (an endogenous ribonucleic acid) identified in human T cells can regulate NFAT, a Ca2+-activated transcription factor, and IL-2 expression in activated T cells, as the synthesis of IL-2 requires NFAT1.
T cells are at rest, NFAT is present in the cytoplasm, while NFAT is phosphorylated and transferred to the nucleus after T cells are activated. NRON interferes with nuclear transport of NFAT (rather than transcriptional activity) by interacting with members of the importin-beta superfamily, including the nuclear transport factor KPNB1 (9). Subsequent studies have shown that NRON acts as a structural scaffold lncRNA where NFAT interacts with GTPase activating protein (IQGAP) and several other NFAT kinase inhibitors (CKE, GSK-β, aDYRK, DYRK), leaving NFAT in the cytoplasm (10). Although these studies give the different regulation patterns, NRON plays a key role on regulating NFAT-dependent IL-2 expression in T cells.
During the differentiation from CD4+ T cells into Th1 and Th2 cells, lincRNAs in T cell subsets can regulate the expression of adjacent protein-coding genes in the upstream promoter region of the protein-coding genes by transcription. As the Th2-specific lincRNA, lincR-Ccr2-5'AS located upstream of the chemokine receptor Ccr2 gene and transcribed in the AS direction, controls the expression of immune genes in Th2 cells together with Gata3. lincR-Ccr2-5'AS also controls the migration of Th2 cells to the lungs in vivo, possibly by controlling the expression of many chemokine receptors (Ccr1, Ccr3, Ccr2 and Ccr5), both of which are located in the same genomic locus as lincR-Ccr2-5'AS (11). However, how lincR-Ccr2-5'AS regulates the expression of these genes remains unclear. In Th2 cells, Th2-LCR lncRNA can regulate the transcription of Th2 cytokines (IL-4, IL-5 and IL-13) genes. Another lincRNA expressed in T cells, growth arrest-specific transcript 5 (GAS5), is associated with cell cycle arrest in mammals that are short of nutrients or exposed to rapamycin (mTOR) antagonists (30-32).
LncRNAs are also expressed in B cells. AS lncRNA (Fas-AS1) controls the production of soluble Fas receptor (sFas), which binds to Fas ligand to regulate Fas-induced apoptosis in B cell lymphoma (33). Fas-AS1 binds to the splicing factor RBM5, that Inhibits RBM5 to skips FAS exon 6 (also known as CD95; TNFRSF6) selectivity, which is necessary for the production of sFas mRNA. Since serum sFas level is associated with poor prognosis of non-Hodgkin’s lymphoma (34), Fas-AS1 has been a potential therapeutic target. In addition, a broad AS interval transcription occurs in the variable (V) region of the immunoglobulin heavy chain (IgH) site in B cells, that is potentially associated with chromatin remodeling, which is related to the diversity of antigenic receptors in developing B-cells (35,36). Whether lncRNAs play a role on maturation and effector function in B cells remains unclear. However, in general, these studies indicated that immune cells expressed a large number of lncRNAs, many of which play a key role on immune response in the host.
At present, it seems that the role of most immune-related lncRNAs is mediated through binding to proteins.
Targets include the splicing factor proline/glutamine-rich (SFPQ) (37), importin-b family (9) and transcription factors, NF-κB (22,23), STAT3 (15), and glucocorticoid receptor (GR) (30) and so on. LncRNAs have shown some functions that it acted as a “bait” to block protein-DNA binding (SFQR, NF-κB and GR) or as an antagonist to block protein-protein interaction (importin-b and STAT3). The immune-related lncRNAs also interact with the hnRNP family (19,24) and chromatin-modifying complex components, including PRC2 (38), core subunit of mixed lineage leukemia (MLL) methyltransferase complex, WD repeat domain 5 (WDR5) and UTX/JMJD3 demethylase (39). Although the mechanism is not completely understood, it is speculated that lncRNAs may combine proteins as scaffolds or target DNA by base pairing (40).
LncRNA and immune related diseases
LncRNA and inflammatory diseases
Up to date, most of the lncRNA-related studies on the immune system focused on functions in mouse and human primary cells and cell lines. However, the role of lncRNAs in human inflammatory diseases have been paid attention. For examples, the expression of lncRNA Morrbid is significantly up-regulated in eosinophils in patients with HES, suggesting that the Morrbid-BCL2L11 axis may be associated with this disease (28). Lnc13 is a highly expressed lncRNA in the bowel of healthy humans, which is significantly down-regulated in patients with chronic diarrheal disease, and inhibits the expression of genes related to inflammatory diseases, suggesting that dysregulated lnc13 may be involved in the inflammatory response of this disease (41). In addition, lnc3 can down-regulate LPS, and may also be an inhibitor of inflammatory response genes (such as Myd88, Stat3 and II1ra). Recently, studies have also found that lincRNA-EPS exposed to microbial environment was inhibited in macrophages and could act as a transcriptional inhibitor to suppress the transcription of immune response gene (IRGs), while the inflammatory response was enhanced in lincRNA-EPS-deficient mice. Moreover, lncRNA-EPS is associated with chromatin and interacts with hnRNPL, a member of the heterologous ribonucleoprotein family, to alter nucleosome localization and inhibit IRGs (42). All of above suggested the existence of some inflammatory suppressive functions of lncRNA. Also, it has been confirmed that linc-MAF-4 was abnormally expressed in multiple sclerosis, and the mutation of lncRNA RMRP was associated with the immunodeficient metaphyseal chondrodysplasia, Mckusick type (CHH) (43,44). LncRNA studies in the immune system indicated that these molecules played a key role in inducing or inhibiting inflammatory diseases.
LncRNA and virus infection
It is documented that many lncRNAs are specifically expressed in immune responses. These lncRNAs showed some important modes to induce the immune responses. Host-pathogen interaction is another important research direction, expect for the study of lncRNAs in the host. LncRNAs have also been involved in the functions of pathogen, which affect the host by a variety of ways. An HIV-1 encoded AS lncRNA may be involved in the establishment of viral latency (45). This AS lncRNA is capable of recruiting epigenetic remodeling complexes containing DNMT3A, a deoxyribonucleic acid methyltransferase, which is involved in epigenetic silencing and RNA-directed transcriptional gene silencing (46).
Another lncRNA is polyadenylated nuclear RNA (PAN) derived from Kaposi’s sarcoma-associated herpesvirus. Like HIV-1 encoded AS lncRNA, PAN is also localized to its own genome. However, PAN is usually responsible for the activation of Kaposi’s sarcoma-associated herpesvirus transcription. If PAN is inhibited, almost all Kaposi’s sarcoma-associated herpesvirus transcription will be blocked. Interestingly, PAN also plays a role in the host gene. Like many of the lncRNAs discussed above, PAN can bind to PRC2 (especially EZH2 and Suz12) to induce heterochromatin formation, which is associated with the downregulation of interferon, IL-18, alpha interferon-16 and RNase L gene (47).
PAN is not the first viral lncRNA to interact with its host: human cytomegalovirus-encoded non-coding transcript, called β2.7 for 2.7 kb in length, was discovered earlier. It has been found that β2.7 interacted with gene associated with retinoid-interferon-induced mortality 19 (GRIM-19) to maintain mitochondrial integrity and function, allowing the virus to complete its life cycle (48).
Similarly, subgenomic flavivirus RNA (sfRNA), produced by West Nile virus, dengue fever, yellow fever, tick-borne encephalitis virus and Japanese encephalitis, is also virus lncRNA. sfRNAs, a class of viral transcripts for about 300–500 bp in length, were first discovered in the Murray Valley virus (49). sfRNAs do not only prevent XRN1 from degradation, but also block XRN1, resulting the accumulation of non-cap structure but stable cellular mRNA, which induces the dysregulation of overall gene expression in cells, and prevent other antiviral RNA from degradation (50). Deletion of sfRNA reduces plaque formation and replication, reflecting the importance of sfRNAs in viral cytopathic and pathogenicity (51). In addition, the replication capacity of West Nile virus without sfRNA is diminished, but it can be obtained again by knockout of interferon-related genes (52). This is a unique regulatory mechanism for lncRNA, suggesting that many other lncRNAs (from viruses or others) have many unknown functions.
LncRNA related to immunity and pregnancy
Pregnancy, an extremely complex and coordinated physiological processes, is considered as a successful allogeneic transplantation and one of the classic models of immune tolerance. Maternal-fetal interface is a key to maintain pregnancy, which can secrete hormones into the maternal circulation to adapt to pregnancy, and transfer maternal antibodies into the fetus except nutrient exchange. Changes of immune environment in the maternal-fetal interface could lead to pregnancy complications such as abortion, preeclampsia and intrauterine growth restriction (53).
Immunotherapy for unexplained recurrent spontaneous abortion (URSA) is effective in clinical, but the mechanism is unclear. The relationship between lncRNAs and URSA has been paid more and more attention. So far, only few studies have identified the relationship between lncRNAs and abortion. One is that lncRNA expression profiles in early recurrent abortion and normal abortion were analyzed, and found that there were 1,449 abnormally expressed lncRNAs, of which 11 up-regulated and 15 down-regulated lncRNAs were involved in biological regulation processes, including mechanisms related to endocrine, immune, extracellular matrix (ECM) receptor interactions, and apoptosis (54). Huang sequenced the transcriptome of URSA villi and found that lnc-SLC4A1-1 was highly expressed in the sample, and could alter trophoblast function by activating immune response via the axis of NF-κB/CXCL8 (55). Another study found that the expression level of lncRNA MEG3 in villi of artificial abortion is lower than that of spontaneous abortion (56).
Endometrial decidualization is also an important part of pregnancy. Hong Wang detected the expression profile and analyzed the function of lncRNAs in gestational sac and decidua of spontaneous abortion, there were more significantly up-regulated lncRNAs associated with infection and inflammation compared with artificial abortion, and there were more lncRNAs in decidua than that in blastocyst (57). Some studies have shown that LINC00473 might mediate the decidualization of human endometrial stromal cells, and the mechanism could be that cAMP-PKA pathway induces STAT3 phosphorylation through IL-11 (58). Although the mechanism is unclear, it is speculated that maternal factor is more significant for the pathogenesis of inflammation-related spontaneous abortion.
The lncRNAs related to preeclampsia include LOC391533, LOC284100, CEACAMP8 (59), SPRY4-IT1 and MALAT1 and so on. SPRY4-IT1 may be involved in trophoblast formation (60), and MALAT1 may be associated with decreased trophoblastic invasion (61). Preeclampsia is associated with abnormal mesenchymal stem cells (MSCs) in maternal-fetal interface. It is interesting that overexpressing MALAT1 in MSCs could promote the polarization of M2 macrophages by inducing IDO expression, suggesting that MALAT1 could improve the immunosuppressive ability of MSCs (62).
The relationship between lncRNAs and fetal growth restriction (FGR) was studied rarely. It has been reported that the expression of lncRNA H19 in placenta of FGR patients was decreased significantly so as to repressed the migration and invasion of extravillous trophoblasts (EVT), which could be related to the decrease of the expression of transforming growth factor beta receptor III (TGFβR3) (63). Up to date, there are few studies about the relationship between immune-related lncRNA and pregnancy. So, whether there is immune-related lncRNA involved in the maintenance of pregnancy and how to maintain pregnancy is still needed to explore in future.
Conclusions
More and more inflammatory diseases are looking for a relationship with lncRNA. Until now, lncRNA has been confirmed that there was a different expression in diseases such as inflammatory bowel disease, diabetes, allergies, asthma and cancer. However, the function of lncRNA in diseases related to inflammation, still needs to be explored. In future, further studies about the relationship between lncRNA and disease will be conducted to provide new ideas for treatment of immune-related diseases.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Award Number: 81771618, recipient: Jing Yang).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Euskirchen GM, Rozowsky JS, Wei CL, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res 2007;17:898-909. [Crossref] [PubMed]
- Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet 2014;30:439-52. [Crossref] [PubMed]
- Liu G, Mattick JS, Taft RJ. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle 2013;12:2061-72. [Crossref] [PubMed]
- Clark MB, Choudhary A, Smith MA, et al. The dark matter rises: the expanding world of regulatory RNAs. Essays Biochem 2013;54:1-16. [Crossref] [PubMed]
- Mattick JS, Amaral PP, Dinger ME, et al. RNA regulation of epigenetic processes. Bioessays 2009;31:51-9. [Crossref] [PubMed]
- Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011;477:295-300. [Crossref] [PubMed]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26-46. [Crossref] [PubMed]
- Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity 2015;42:792-804. [Crossref] [PubMed]
- Willingham AT, Orth AP, Batalov S, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005;309:1570-3. [Crossref] [PubMed]
- Sharma S, Findlay GM, Bandukwala HS, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A 2011;108:11381-6. [Crossref] [PubMed]
- Hu G, Tang Q, Sharma S, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol 2013;14:1190-8. [Crossref] [PubMed]
- Collier SP, Collins PL, Williams CL, et al. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol 2012;189:2084-8. [Crossref] [PubMed]
- Collier SP, Henderson MA, Tossberg JT, et al. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J Immunol 2014;193:3959-65. [Crossref] [PubMed]
- Ranzani V, Rossetti G, Panzeri I, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol 2015;16:318-25. [Crossref] [PubMed]
- Wang P, Xue Y, Han Y, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014;344:310-3. [Crossref] [PubMed]
- Zhang X, Lian Z, Padden C, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009;113:2526-34. [Crossref] [PubMed]
- Pang KC, Dinger ME, Mercer TR, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol 2009;182:7738-48. [Crossref] [PubMed]
- Xia F, Dong F, Yang Y, et al. Dynamic transcription of long non-coding RNA genes during CD4+ T cell development and activation. PLoS One 2014;9:e101588. [Crossref] [PubMed]
- Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013;341:789-92. [Crossref] [PubMed]
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223-7. [Crossref] [PubMed]
- Carpenter S. Determining the Function of Long Noncoding RNA in Innate Immunity. Methods Mol Biol 2016;1390:183-95. [Crossref] [PubMed]
- Rapicavoli NA, Qu K, Zhang J, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2013;2:e00762. [Crossref] [PubMed]
- Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife 2014;3:e01776. [Crossref] [PubMed]
- Li Z, Chao TC, Chang KY, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A 2014;111:1002-7. [Crossref] [PubMed]
- Hirose T, Virnicchi G, Tanigawa A, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 2014;25:169-83. [Crossref] [PubMed]
- Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med 2014;20:623-31. [Crossref] [PubMed]
- Wei S, Zhao M, Wang X, et al. PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J Hematol Oncol 2016;9:44. [Crossref] [PubMed]
- Kotzin JJ, Spencer SP, McCright SJ, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 2016;537:239-43. [Crossref] [PubMed]
- Liu AY, Torchia BS, Migeon BR, et al. The human NTT gene: identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics 1997;39:171-84. [Crossref] [PubMed]
- Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010;3:ra8. [Crossref] [PubMed]
- Mourtada-Maarabouni M, Hasan AM, Farzaneh F, et al. Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5). Mol Pharmacol 2010;78:19-28. [Crossref] [PubMed]
- Mourtada-Maarabouni M, Hedge VL, Kirkham L, et al. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5). J Cell Sci 2008;121:939-46. [Crossref] [PubMed]
- Sehgal L, Mathur R, Braun FK, et al. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia 2014;28:2376-87. [Crossref] [PubMed]
- Niitsu N, Sasaki K, Umeda M. A high serum soluble Fas/APO-1 level is associated with a poor outcome of aggressive non-Hodgkin's lymphoma. Leukemia 1999;13:1434-40. [Crossref] [PubMed]
- Verma-Gaur J, Torkamani A, Schaffer L, et al. Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci U S A 2012;109:17004-9. [Crossref] [PubMed]
- Bolland DJ, Wood AL, Johnston CM, et al. Antisense intergenic transcription in V(D)J recombination. Nat Immunol 2004;5:630-7. [Crossref] [PubMed]
- Imamura K, Akimitsu N. Long Non-Coding RNAs Involved in Immune Responses. Front Immunol 2014;5:573. [Crossref] [PubMed]
- Rossetto CC, Tarrant-Elorza M, Pari GS. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog 2013;9:e1003366. [Crossref] [PubMed]
- Rossetto CC, Pari G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog 2012;8:e1002680. [Crossref] [PubMed]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013;14:699-712. [Crossref] [PubMed]
- Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, et al. A long noncoding RNA associated with susceptibility to celiac disease. Science 2016;352:91-5. [Crossref] [PubMed]
- Atianand MK, Hu W, Satpathy AT, et al. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell 2016;165:1672-85. [Crossref] [PubMed]
- Huang W, Thomas B, Flynn RA, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 2015;528:517-22. [Crossref] [PubMed]
- Zhang F, Liu G, Wei C, et al. Linc-MAF-4 regulates Th1/Th2 differentiation and is associated with the pathogenesis of multiple sclerosis by targeting MAF. FASEB J 2017;31:519-25. [Crossref] [PubMed]
- Saayman S, Ackley A, Turner AW, et al. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther 2014;22:1164-75. [Crossref] [PubMed]
- Weinberg MS, Villeneuve LM, Ehsani A, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 2006;12:256-62. [Crossref] [PubMed]
- Rossetto CC, Tarrant-Elorza M, Verma S, et al. Regulation of viral and cellular gene expression by Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA. J Virol 2013;87:5540-53. [Crossref] [PubMed]
- Reeves MB, Davies AA, McSharry BP, et al. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 2007;316:1345-8. [Crossref] [PubMed]
- Urosevic N, van Maanen M, Mansfield JP, et al. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. J Gen Virol 1997;78:23-9. [Crossref] [PubMed]
- Moon SL, Anderson JR, Kumagai Y, et al. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 2012;18:2029-40. [Crossref] [PubMed]
- Pijlman GP, Funk A, Kondratieva N, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008;4:579-91. [Crossref] [PubMed]
- Schuessler A, Funk A, Lazear HM, et al. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J Virol 2012;86:5708-18. [Crossref] [PubMed]
- Roberts CT. IFPA Award in Placentology Lecture: Complicated interactions between genes and the environment in placentation, pregnancy outcome and long term health. Placenta 2010;31 Suppl:S47-53. [Crossref] [PubMed]
- Wang L, Tang H, Xiong Y, et al. Differential expression profile of long noncoding RNAs in human chorionic villi of early recurrent miscarriage. Clin Chim Acta 2017;464:17-23. [Crossref] [PubMed]
- Huang Z, Du G, Huang X, et al. The enhancer RNA lnc-SLC4A1-1 epigenetically regulates unexplained recurrent pregnancy loss (URPL) by activating CXCL8 and NF-kB pathway. EBioMedicine 2018;38:162-70. [Crossref] [PubMed]
- Ankolkar M, Salvi V, Warke H, et al. Methylation status of imprinted genes DLK1-GTL2, MEST (PEG1), ZAC (PLAGL1), and LINE-1 elements in spermatozoa of normozoospermic men, unlike H19 imprinting control regions, is not associated with idiopathic recurrent spontaneous miscarriages. Fertil Steril 2013;99:1668-73. [Crossref] [PubMed]
- Wang H, Cao Q, Ge J, et al. LncRNA-regulated infection and inflammation pathways associated with pregnancy loss: genome wide differential expression of lncRNAs in early spontaneous abortion. Am J Reprod Immunol 2014;72:359-75. [Crossref] [PubMed]
- Liang XH, Deng WB, Liu YF, et al. Non-coding RNA LINC00473 mediates decidualization of human endometrial stromal cells in response to cAMP signaling. Sci Rep 2016;6:22744. [Crossref] [PubMed]
- He X, He Y, Xi B, et al. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS One 2013;8:e81437. [Crossref] [PubMed]
- Zou Y, Jiang Z, Yu X, et al. Upregulation of long noncoding RNA SPRY4-IT1 modulates proliferation, migration, apoptosis, and network formation in trophoblast cells HTR-8SV/neo. PLoS One 2013;8:e79598. [Crossref] [PubMed]
- Chen H, Meng T, Liu X, et al. Long non-coding RNA MALAT-1 is downregulated in preeclampsia and regulates proliferation, apoptosis, migration and invasion of JEG-3 trophoblast cells. Int J Clin Exp Pathol 2015;8:12718-27. [PubMed]
- Li X, Song Y, Liu F, et al. Long Non-Coding RNA MALAT1 Promotes Proliferation, Angiogenesis, and Immunosuppressive Properties of Mesenchymal Stem Cells by Inducing VEGF and IDO. J Cell Biochem 2017;118:2780-91. [Crossref] [PubMed]
- Zuckerwise L, Li J, Lu L, et al. H19 long noncoding RNA alters trophoblast cell migration and invasion by regulating TbetaR3 in placentae with fetal growth restriction. Oncotarget 2016;7:38398-407. [Crossref] [PubMed]


