The role of CYP3A4 in the biotransformation of bile acids and therapeutic implication for cholestasis
Introduction
Cytochrome P450s (CYPs) are a superfamily of enzymes, which catalyse a broad range of compounds (1,2). These enzymes detoxify xenobiotics to exert body defence against environmental hazard. They also play key roles in carcinogenesis by turning precarcinogens into carcinogen such as azoxymethane and deactivating carcinogens (3). CYPs also regulate many human body biological processes such as biosynthesis of estrogen, conversion of cholesterol into bile acids and biotransformation of bile acids (4). Manipulation of these processes through CYP activities may help prevention and treatment of diseases. In this review, we summarise the roles of CYP3A4 in biotransformation of bile acids, regulation of CYP3A4 and stimulation of CYP3A4 activity for the treatment of cholestasis.
Bile acids are the major products of cholesterol catabolism, which have important physiological functions in human body. They facilitate not only the elimination of cholesterol and body wastes but also the digestion of fat and the absorption of lipid soluble vitamins. Bile acids also play a role as signalling molecules in the fine-tuning of energy homeostasis (5,6). However, bile acids are toxic when accumulated in high concentration. Cholestasis is the impairment of bile flow and is characterised by the accumulation of bile acids in the liver and serum (7). The accumulated bile acids are toxic and critical in the pathophysiology of cholestasis. A major aim of cholestasis treatment is to reduce the concentration of toxic bile acids in both blood and liver.
The toxicity of accumulated bile acids has been shown both in vivo and in vitro, which result in a series of pathological changes including alterations of P450 activity. In a 5 day bile duct ligated model, total P450 was decreased to 50% and 75% of controls in male and female rats respectively (7,8). In vitro, addition of bile acids dramatically inhibited P450-catalysed reactions (9). The effect correlates with the hydrophobicity of bile acids with most toxic being the lithocholic acids, the moderately toxic deoxycholic and chenodeoxycholic acids and the least toxic cholic acid. At a cellular level, bile acids caused apoptosis at low concentrations and necrosis at high concentrations.
There are adaptive responses in the body to decrease the pool of bile acids to protect the body from their toxicity. These responses include decreased synthesis, increased metabolism and renal elimination of bile acids. Nuclear receptors have the important functional roles in these responses and hold much promise in the treatment of cholestasis (10). CYP3A4 is the major enzyme for the detoxification of bile acids, hydrolysing bile acids to facilitate their subsequent glucuronidation by UDP-glucuronosyltransferases (UGTs) (11). Stimulation of CYP3A4 activity could be employed for the treatment of cholestasis.
Role of CYP3A4 and its regulation
CYP3A4 is a member of cytochrome P450 (CYP450) enzymes encoded by a gene superfamily (12). Among all CYP enzymes, CYP3A4 is the most important, which is the most abundant in both liver and intestinal tract (13). The gene encoding CYP3A4 is located in chromosome 7q21.1. Other CYP enzymes encoded in 7q21.1 are CYP3A5, CYP3A7 and CYP3A43. But they are minor isoforms.
The enzyme CYP3A4 has broad substrates (Table 1). The broad substrates are attributed to the structure of the CYP3A4 active site which has been characterised by X-ray crystallography (23). It has been reported that CYP3A4 has a much larger active site cavity to bind a broad range of substrates, compared with other CYPs (23,24). CYP3A4 also has multiple binding sites (25). The substrates are generally transformed into inactive forms or compounds of lower activity through reactions such as N-oxidation, C-oxidation, N-dealkylation, O-dealkylation, nitro-reduction, dehydration and C-hydroxylation. CYP3A4 metabolises not only xenobiotics including majority of drugs and carcinogens to protect living organisms from environmental toxicity (12,26) but also many endogenous compounds such as cholesterol, bile acids, fatty acids, prostaglandins, leukotrienes, retinoids and biogenic amines (12,27).

Full table
CYP3A4 expression, its stability and inhibition are regulated by different mechanisms. Many factors including nuclear receptors, hormones, xenobiotics and signalling molecules regulate CYP3A4 expression as shown in Figure 1 (27,28). In CYP3A4 regulatory region, there are two binding sites for nuclear receptors PXR and FXR to bind to and confer transactivation (29). PXR forms a heterodimer with 9-cis retinoic acid receptor (RXR) to bind to CYP3A4 promoters (14,30,31). Disruption of these elements in the promoter of CYP3A4 caused dramatic decrease in reporter activities (30). CYP3A4 dis-regulation in PXR-null mice further demonstrated an important role of PXR in CYP3A4 activation (32). Another nuclear receptor constitutive androstane receptor (CAR) has also been demonstrated to activate CYP3A4 (33,34). Many hormones such as triiodothyronine, dexamethasone and growth hormone involve the regulation of CYP3A4 (20). CYP3A4 is also regulated by a large number of xenobiotics including many drugs (Table 1) (18,21,31,35). Several drugs, some beverage and bile acids can inhibit CYP3A4 activity (9,15,22,36-38).
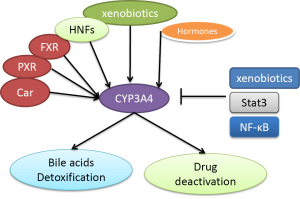
Several signalling molecules have also been demonstrated to regulate CYP3A4. These molecules include C/EBPa and C/EBPb, HNF1a and HNF3gamma, HNF4a, USF, AMPK and PPAR-gamma (39-43). Jover et al. showed that JAK/Stat pathway down-regulates CYP3A4 (39). NF-κB was also shown to decrease CYP3A stability (44).
CYP3A4 in bile acids transformation and its alterations in cholestasis
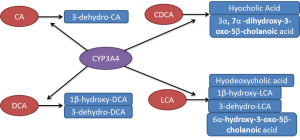
CYP3A4 also plays an important role in the detoxification of bile acids where it catalyses their hydroxylation thereby increasing the hydrophility of bile acids and thus decreasing their toxicity. Bile acids are synthesised from cholesterol and rate-limiting enzyme is CYP7A1. It is subject to feedback inhibition by bile acids. The bile acids are also metabolised by CYP3A4 (45,46), which exerts an essential protective effect in cholestasis. Several studies have characterised the products formed by CYP3A4 from different bile acids (Figure 2). Chenodeoxycholic acid is turned into both hyocholic acid (3α, 6α, 7α-trihydroxy-5β-cholanoic acid) and 3α, 7α-dihydroxy-3-oxo-5β-cholanoic acid while only one product is formed from cholic acid, 3-dehydro-CA (7α, 12α-dihydroxy-3-oxo-5β-cholanoic acid) (47,48). Deoxycholic acid (DCA) is catalysed into 3-dehydro-DCA (12α-hydroxy-3-oxo-5β-cholanoic acid) and 1β, 3α, 12α-trihydroxy-5β-cholanoic acid (47,48). Lithocholic acid was metabolised into four products 3-oxo-5β-cholanoic acid (3-dehydro-LCA), 6α-hydroxy-3-oxo-5β-cholanoic acid, 3α, 6α-dihydroxy-5β-cholanoic acid (hyodeoxycholic acid) and 1β, 3α-dihydroxy-5β-cholanoic acid (1β-hydroxy-LCA) (47,49,50). Patients treated with the antiepileptic drug carbamazepine, a CYP3A4 inducer, markedly elevated urinary excretion of 1β-hydroxydeoxycholic acid. Both taurochenodeoxycholic acid and lithocholic acid have been subjected to metabolism by fourteen recombinant expressed CYPs (51). Actually, only CYP3A4 was able to catalyse 6-α-hydroxylation of these two bile acids. The Vmax and Km for 6-α-hydroxylation of taurochenodeoxycholic acid catalysed by CYP3A4 was 18.2 nmol/nmol P450/min and 90 µM, respectively. Furthermore, 6-α-hydroxylation of these two bile acids has been found to correlate with the levels of CYP3A4 and 6-β-hydroxylation of testosterone in ten human liver microsomes (51). These reactions are completely inhibited by 10 µM troleandomycin, a specific inhibitor of CYP3A4 (51).
It has been shown that CYP3A4 is distinctly increased in several cholestasis models and also upregulated in bile duct-ligated rats (BDL) (52). This could facilitate detoxification of bile acids and increase their renal elimination. However, increased bile acids have detergent effect, which decreases the CYP3A4 activity (9,53).
Changed bile acid pool size by CYP3A4
Both liver and kidney are the two organs involved in the excretion of toxic substrates with each demonstrating complementary effects. In cholestasis, the major route for bile acid secretion through liver is blocked and thus, excretion by the kidney is greatly increased. In cholestasis patients, there was increased urinary excretion of 6α-hydroxylated bile acids, a-hyocholic acid and hyodeoxycholic acid (54-56). In cholestasis of pregnancy, there was a ten- to one hundred-fold increase in cholic and chenodeoxycholic acid in the urine while the amounts of 1- and 6-hydroxylated products of cholic acid were also markedly increased (57).
In bile duct ligated rats, urinary excretion of bile acids was increased by more than 380-fold from 0.05 to 19 µmol/day (58). The urinary excretion of 6β-hydroxylated bile acids and β-muricholic acid also increased (16,58,59). The ratio of cholic acid/β-muricholic acid in urine changed from 5:3 at day 1 to 1:8 at day 5 (16). This indicates metabolism of bile acids by CYP3A4 is increased in cholestasis and bile acids are detoxified and easier to be excluded through urinary.
Up-regulation of CYP3A4 expression in cholestasis
The induction of CYP3A4 expression by bile acids is mediated by FXR (30,60) and PXR (52,60,61). It has been reported that chenodeoxycholic acid enhance CYP3A4 expression via FXR (29). The regulatory sequences have recently been identified (62). There are two functional FXR recognition sites located in a 345 bp element within the regulatory region of CYP3A4 (29).
PXR forms a heterodimer with retinoid X receptor (RXR) to bind to xenobiotic response elements in the CYP3A4 promoter (63-66). Disruption of the PXR gene eliminated the enhanced expression of CYP3A4 by its inducers dexamethasone and pregnenolone 16a-carbonitrile (32). In PXR-null mice, introduction of human PXR allowed the induction of CYP3A4 by the human-specific inducer-rifampicin (32). In the regulatory region of CYP3A4 gene, two PXR elements (distant and proximal) have been identified that are separated by more than 7,000 nucleotides in the genome (30). Disruption of the distant and proximal elements causes a 73% and 53% decrease in reporter activities respectively. However, in vitro binding studies showed that the proximal element was preferred. Chromatin immunoprecipitation experiments showed that the genomic fragment harbouring the proximal element was preferably precipitated over the fragment containing the distant element in the CYP3A4 gene (67).
Bile acids were demonstrated to regulate PXR in an in vitro assay employing a chimeric reporter system in which the FXR ligand-binding domain was fused with a reporter gene. Addition of lithocholic acid, deoxycholic acid and chenodeoxycholic acid activated the reporter construct 5 fold (68). This was confirmed by the treatment of mice with lithocholic acid, 4 days of lithocholic acid administration caused liver necrosis in 50% of wild mice increasing to 100% in PXR-deficient mice (32). Another study demonstrated that PXR was upregulated by lithocholic acid and its 3-keto metabolite (19).
Other nuclear receptors such as CAR may be also involved in the regulation of CYP3A4 (69). CAR has been demonstrated to regulate CYP3A4 transcription (30). It has a crosstalk with PXR to regulate overlapping but distinct genes. However, so far, no evidence supports the binding of bile acids to CAR.
Bile acids have been also shown to bind to the vitamin D receptor with low affinity to activate CYP3A4 (70-73). Lithocholic acid was shown to activate CYP3A4 mainly via the vitamin D receptor in colon, where PXR appeared not involved as Dex failed to stimulate CYP3A4 expression. Other bile acids appear not to bind to this receptor. However, a lithocholic acid derivative—lithocholic acid acetate was 30 times more efficient at activating CYP3A4 than lithocholic acid (72). The binding model of lithocholic acid is different to that of 1,25-dihydroxyvitamin D3 which has a higher receptor affinity. Thus, two vitamin D receptor elements with distinctly different DNA motifs were identified in the regulatory region of CYP3A4. This indicates that these two compounds have the complementary roles in CYP3A4 mediated detoxification. However, it remains unclear whether the vitamin D receptor plays a role in the expression of CYP3A4 by bile acids in the liver.
Detergent effect of bile acids on CYP3A4 activity and regulatory role of estrogen
Accumulated bile acids can inhibit CYP3A4 activity. As shown in vitro, they have non-specific detergent effects on several rat CYPs and non-CYPs (9). It is quite possible that CYP3A4 is also subjected to the direct inhibition of accumulated bile acids. This should be taken into account when considering the actual activity of CYP3A4 in cholestasis.
Estrogen has an important role in cholestasis. It could cause cholestasis especially in patients with bile acid transporter deficiency. It has been found to accumulate in BDL rats (7,8) and to down-regulate male specific CYPs 2C11 and 3A2. Recently, estrogen was considered to down-regulate CYP3A4 in human liver. After comparing 27 human hepatic CYP3A4 mRNA levels, it was found that young females have lower levels of CYP3A4 mRNA than young men, while older women displayed the same levels of CYP3A4 as older men, both of which were much higher than those in young women (74). Thus, the implication is that estrogen may play a role in regulating CYP3A4 transcription. However, another study using 54 samples of human liver microsomes did not find any influence of age or gender on CYP3A4-mediated midazolam hydroxylation and CYP3A4 protein mass (75).
Overall, activity of CYP3A4 will be determined by (I) transcriptional up-regulation of CYP3A4 expression by bile acids through nuclear receptors, (II) down-regulation by blood levels of estrogen and (III) direct inhibition of CYP3A4 by bile acids due to detergent effect of bile acids. Therefore, CYP3A4 activity may be stimulated at the early stage of cholestasis but inhibited when cholestasis further develops due to highly increased blood levels of estrogen and bile acids.
Polymorphism in CYP3A4 mediated bile acids detoxification
CYP3A4 expression is highly variable. There are marked individual variations in the expression of CYPs including CYP3A4, which affect the biological response (efficacy and/or toxicity) of many drugs as well as individual responses to CYP inducers (76,77). In humans, a 10-fold variation exists among individuals for rifampicin induced CYP3A4 expression (78-80). In the liver, substantial differences in CYP3A4 expression between individuals have been detected that could not be explained by environmental factors (78). A similar situation was found in the intestine, whereby CYP3A4 mRNA levels in biopsies of the mucosa of five healthy subjects varied by about 10 fold (79). This variation could be related to the polymorphisms of CYP3A4 and the other genes involved in the regulation of CYP3A4.
Any polymorphism in the genes involved in CYP3A4 mediated bile acid detoxification in cholestasis could affect this adaptive response. More than 20 CYP3A4 mutations have been identified in humans although the frequency is low (81). Mutations of CYP3A4 affect the metabolism of clinically used drugs, leading to toxic blood concentration of drugs. Theoretically, these CYP3A4 variations can also affect bile acid detoxification. However, there have been no reports on them as there have been for bile acids transporters. There is also a report that a high degree of SNP polymorphism in PXR found in a screening of the Dutch population (66). How these mutations affect the cholestasis is not yet assessed.
Therapeutic implications
The nuclear receptor activators have been extensively studied and some of them are actually used clinically. Rifampicin, for example, is used for treatment of cholestasis (82-86). In 14 healthy volunteers, administration of rifampicin at 600 mg for 7 days markedly increased urinary excretion of bile acids, the 6-α-glucuronides, caused by increased production of 6 hydroxylated hyocholic and hyodeoxycholic acids, with subsequently conjugation at C-6 by UDP-glucuronosyltransferase (87).
Ursodeoxycholic acid (UDCA) is used extensively although SW4064 and 6ECDCA have been shown to be much more potent inducers (86). UDCA has been used clinically for many years in the treatment of cholestasis. Although it was shown to have beneficial effect on the liver with cholestasis, the effect was mild (10). More effective compounds could be tested to improve the treatment of cholestasis.
As shown in Figure 1 and Table 1, CYP3A4 activity is stimulated by many other compounds. These CYP3A4 activators may also be considered for use in treating cholestasis. There are possibilities to discover a better compound than UDCA and rifampicin in terms of effectiveness in reducing the levels of bile acids both in liver and blood. It is also possible to use some of them for combinatorial application with rifampicin or UDCA. At present these stimulators are not well studied in the treatment of cholestasis.
CYP3A4 catalyzes more than 50% of clinically used drugs. Any change of CYP3A4 activity will affect the pharmacokinetics of these drugs. Thus, the manipulation of CYP3A4 in cholestasis must be incorporated into the consideration of the dose of other drugs used in these patients. It also affects the metabolism of other drugs such as anti-cancer drug cyclophosphamide (17), cardiovascular drug nifedipine (88). The main drugs affected are listed in Table 1.
Conclusions
CYP3A4 is an important enzyme in bile acid detoxification, which is responsively up-regulated in cholestasis. Disruption of the CYP adaptive responses in cholestasis leads to more severe hepatic injury, thus retaining of high levels of CYP3A4 protein and its enzymatic activity by CYP3A4 inducers has therapeutic implication. Clinically, it is essential, to reduce the factors which inhibit CYP3A4 and to employ CYP3A4 activators to maximize its activity for detoxification of bile acids in the treatment of cholestasis.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Zhang T, Zhao M, Pang Y, et al. Recent progress on bioinformatics, functional genomics, and metabolomics research of cytochrome P450 and its impact on drug discovery. Curr Top Med Chem 2012;12:1346-55. [PubMed]
- Kumar S, Sharma R, Roychowdhury A. Modulation of cytochrome-P450 inhibition (CYP) in drug discovery: a medicinal chemistry perspective. Curr Med Chem 2012;19:3605-21. [PubMed]
- Chen J, Huang XF. The signal pathways in azoxymethane-induced colon cancer and preventive implications. Cancer Biol Ther 2009;8:1313-7. [PubMed]
- McLean KJ, Hans M, Munro AW. Cholesterol, an essential molecule: diverse roles involving cytochrome P450 enzymes. Biochem Soc Trans 2012;40:587-93. [PubMed]
- Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J 2006;25:1419-25. [PubMed]
- Watanabe M, Houten SM, Mataki C, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006;439:484-9. [PubMed]
- Chen J, Murray M, Liddle C, et al. Downregulation of male-specific cytochrome P450s 2C11 and 3A2 in bile duct-ligated male rats: importance to reduced hepatic content of cytochrome P450 in cholestasis. Hepatology 1995;22:580-7. [PubMed]
- Chen J, Robertson G, Field J, et al. Effects of bile duct ligation on hepatic expression of female-specific CYP2C12 in male and female rats. Hepatology 1998;28:624-30. [PubMed]
- Chen J, Farrell GC. Bile acids produce a generalized reduction of the catalytic activity of cytochromes P450 and other hepatic microsomal enzymes in vitro: relevance to drug metabolism in experimental cholestasis. J Gastroenterol Hepatol 1996;11:870-7. [PubMed]
- Chen J, Raymond K. Nuclear receptors, bile-acid detoxification, and cholestasis. Lancet 2006;367:454-6. [PubMed]
- Takeda S, Ishii Y, Iwanaga M, et al. Modulation of UDP-glucuronosyltransferase function by cytochrome P450: evidence for the alteration of UGT2B7-catalyzed glucuronidation of morphine by CYP3A4. Mol Pharmacol 2005;67:665-72. [PubMed]
- Nelson DR, Koymans L, Kamataki T, et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 1996;6:1-42. [PubMed]
- Scott EE, Halpert JR. Structures of cytochrome P450 3A4. Trends Biochem Sci 2005;30:5-7. [PubMed]
- Waxman DJ. P450 gene induction by structurally diverse xenochemicals: central role of nuclear receptors CAR, PXR, and PPAR. Arch Biochem Biophys 1999;369:11-23. [PubMed]
- Zhou S, Yung Chan S, Cher Goh B, et al. Mechanism-based inhibition of cytochrome P450 3A4 by therapeutic drugs. Clin Pharmacokinet 2005;44:279-304. [PubMed]
- Takita M, Ikawa S, Ogura Y. Effect of bile duct ligation on bile acid and cholesterol metabolism in rats. J Biochem 1988;103:778-86. [PubMed]
- Zhang J, Tian Q, Yung Chan S, et al. Metabolism and transport of oxazaphosphorines and the clinical implications. Drug Metab Rev 2005;37:611-703. [PubMed]
- Nallani SC, Glauser TA, Hariparsad N, et al. Dose-dependent induction of cytochrome P450 (CYP) 3A4 and activation of pregnane X receptor by topiramate. Epilepsia 2003;44:1521-8. [PubMed]
- Staudinger JL, Goodwin B, Jones SA, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A 2001;98:3369-74. [PubMed]
- Liddle C, Goodwin BJ, George J, et al. Separate and interactive regulation of cytochrome P450 3A4 by triiodothyronine, dexamethasone, and growth hormone in cultured hepatocytes. J Clin Endocrinol Metab 1998;83:2411-6. [PubMed]
- Raucy JL. Regulation of CYP3A4 expression in human hepatocytes by pharmaceuticals and natural products. Drug Metab Dispos 2003;31:533-9. [PubMed]
- Paine MF, Criss AB, Watkins PB. Two major grapefruit juice components differ in intestinal CYP3A4 inhibition kinetic and binding properties. Drug Metab Dispos 2004;32:1146-53. [PubMed]
- Yano JK, Wester MR, Schoch GA, et al. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem 2004;279:38091-4. [PubMed]
- Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci U S A 2006;103:13682-87. [PubMed]
- Houston JB, Galetin A. Progress towards prediction of human pharmacokinetic parameters from in vitro technologies. Drug Metab Rev 2003;35:393-415. [PubMed]
- Shayeganpour A, El-Kadi AO, Brocks DR. Determination of the enzyme(s) involved in the metabolism of amiodarone in liver and intestine of rat: the contribution of cytochrome P450 3A isoforms. Drug Metab Dispos 2006;34:43-50. [PubMed]
- Christians U. Transport proteins and intestinal metabolism: P-glycoprotein and cytochrome P4503A. Ther Drug Monit 2004;26:104-6. [PubMed]
- Savas U, Griffin KJ, Johnson EF. Molecular mechanisms of cytochrome P-450 induction by xenobiotics: An expanded role for nuclear hormone receptors. Mol Pharmacol 1999;56:851-7. [PubMed]
- Gnerre C, Blättler S, Kaufmann MR, et al. Regulation of CYP3A4 by the bile acid receptor FXR: evidence for functional binding sites in the CYP3A4 gene. Pharmacogenetics 2004;14:635-45. [PubMed]
- Goodwin B, Hodgson E, Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 1999;56:1329-39. [PubMed]
- Chen J, Raymond K. Roles of rifampicin in drug-drug interactions: underlying molecular mechanisms involving the nuclear pregnane X receptor. Ann Clin Microbiol Antimicrob 2006;5:3. [PubMed]
- Xie W, Barwick JL, Downes M, et al. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature 2000;406:435-9. [PubMed]
- Burk O, Koch I, Raucy J, et al. The induction of cytochrome P450 3A5 (CYP3A5) in the human liver and intestine is mediated by the xenobiotic sensors pregnane X receptor (PXR) and constitutively activated receptor (CAR). J Biol Chem 2004;279:38379-85. [PubMed]
- Faucette SR, Sueyoshi T, Smith CM, et al. Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther 2006;317:1200-9. [PubMed]
- Hariparsad N, Nallani SC, Sane RS, et al. Induction of CYP3A4 by efavirenz in primary human hepatocytes: comparison with rifampin and phenobarbital. J Clin Pharmacol 2004;44:1273-81. [PubMed]
- Bailey DG, Dresser GK, Bend JR. Bergamottin, lime juice, and red wine as inhibitors of cytochrome P450 3A4 activity: comparison with grapefruit juice. Clin Pharmacol Ther 2003;73:529-37. [PubMed]
- Zhou S, Chan E, Lim LY, et al. Therapeutic drugs that behave as mechanism-based inhibitors of cytochrome P450 3A4. Curr Drug Metab 2004;5:415-42. [PubMed]
- Hidaka M, Okumura M, Fujita K, et al. Effects of pomegranate juice on human cytochrome p450 3A (CYP3A) and carbamazepine pharmacokinetics in rats. Drug Metab Dispos 2005;33:644-8. [PubMed]
- Jover R, Bort R, Gómez-Lechón MJ, et al. Down-regulation of human CYP3A4 by the inflammatory signal interleukin-6: molecular mechanism and transcription factors involved. FASEB J 2002;16:1799-801. [PubMed]
- Rodríguez-Antona C, Bort R, Jover R, et al. Transcriptional regulation of human CYP3A4 basal expression by CCAAT enhancer-binding protein alpha and hepatocyte nuclear factor-3 gamma. Mol Pharmacol 2003;63:1180-9. [PubMed]
- Biggs JS, Wan J, Cutler NS, et al. Transcription factor binding to a putative double E-box motif represses CYP3A4 expression in human lung cells. Mol Pharmacol 2007;72:514-25. [PubMed]
- Tirona RG, Lee W, Leake BF, et al. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med 2003;9:220-4. [PubMed]
- Rencurel F, Foretz M, Kaufmann MR, et al. Stimulation of AMP-activated protein kinase is essential for the induction of drug metabolizing enzymes by phenobarbital in human and mouse liver. Mol Pharmacol 2006;70:1925-34. [PubMed]
- Zangar RC, Bollinger N, Verma S, et al. The nuclear factor-kappa B pathway regulates cytochrome P450 3A4 protein stability. Mol Pharmacol 2008;73:1652-8. [PubMed]
- Bodin K, Lindbom U, Diczfalusy U. Novel pathways of bile acid metabolism involving CYP3A4. Biochim Biophys Acta 2005;1687:84-93.
- Hofmann AF. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab Rev 2004;36:703-22. [PubMed]
- Bodin K, Lindbom U, Diczfalusy U. Novel pathways of bile acid metabolism involving CYP3A4. Biochim Biophys Acta 2005;1687:84-93.
- Gustafsson J, Andersson S, Sjövall J. Bile acid metabolism during development: metabolism of taurodeoxycholic acid in human fetal liver. Biol Neonate 1985;47:26-31. [PubMed]
- Araya Z, Wikvall K. 6alpha-hydroxylation of taurochenodeoxycholic acid and lithocholic acid by CYP3A4 in human liver microsomes. Biochim Biophys Acta 1999;1438:47-54. [PubMed]
- Gustafsson J, Anderson S, Sjövall J. Bile acid metabolism during development: metabolism of lithocholic acid in human fetal liver. Pediatr Res 1987;21:99-103. [PubMed]
- Araya Z, Wikvall K. 6alpha-hydroxylation of taurochenodeoxycholic acid and lithocholic acid by CYP3A4 in human liver microsomes. Biochim Biophys Acta 1999;1438:47-54. [PubMed]
- Stedman C, Robertson G, Coulter S, et al. Feed-forward regulation of bile acid detoxification by CYP3A4: studies in humanized transgenic mice. J Biol Chem 2004;279:11336-43. [PubMed]
- Palmeira CM, Rolo AP. Mitochondrially-mediated toxicity of bile acids. Toxicology 2004;203:1-15. [PubMed]
- Bremmelgaard A, Almé B. Analysis of plasma bile acid profiles in patients with liver diseases associated with cholestasis. Scand J Gastroenterol 1980;15:593-600. [PubMed]
- Almé B, Bremmelgaard A, Sjövall J, et al. Analysis of metabolic profiles of bile acids in urine using a lipophilic anion exchanger and computerized gas-liquid chromatorgaphy-mass spectrometry. J Lipid Res 1977;18:339-62. [PubMed]
- Eklund A, Norlander A, Norman A. Bile acid synthesis and excretion following release of total extrahepatic cholestasis by percutaneous transhepatic drainage. Eur J Clin Invest 1980;10:349-55. [PubMed]
- Thomassen PA. Urinary bile acids in late pregnancy and in recurrent cholestasis of pregnancy. Eur J Clin Invest 1979;9:425-32. [PubMed]
- Dueland S, Reichen J, Everson GT, et al. Regulation of cholesterol and bile acid homoeostasis in bile-obstructed rats. Biochem J 1991;280:373-7. [PubMed]
- Kinugasa T, Uchida K, Kadowaki M, et al. Effect of bile duct ligation on bile acid metabolism in rats. J Lipid Res 1981;22:201-7. [PubMed]
- Zhang J, Huang W, Qatanani M, et al. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem 2004;279:49517-22. [PubMed]
- Bock HH, Lammert F. Nuclear xeno-sensors as receptors for cholestatic bile acids: the second line of defense. Hepatology 2002;35:232-4. [PubMed]
- Martínez-Jiménez CP, Gómez-Lechón MJ, Castell JV, et al. Transcriptional regulation of the human hepatic CYP3A4: identification of a new distal enhancer region responsive to CCAAT/enhancer-binding protein beta isoforms (liver activating protein and liver inhibitory protein). Mol Pharmacol 2005;67:2088-101. [PubMed]
- Lehmann JM, McKee DD, Watson MA, et al. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 1998;102:1016-23. [PubMed]
- Li T, Chiang JY. Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4alpha and coactivators, and suppression of small heterodimer partner gene expression. Drug Metab Dispos 2006;34:756-64. [PubMed]
- Faucette SR, Sueyoshi T, Smith CM, et al. Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther 2006;317:1200-9. [PubMed]
- Gu X, Ke S, Liu D, et al. Role of NF-kappaB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. J Biol Chem 2006;281:17882-9. [PubMed]
- Song X, Xie M, Zhang H, et al. The pregnane X receptor binds to response elements in a genomic context-dependent manner, and PXR activator rifampicin selectively alters the binding among target genes. Drug Metab Dispos 2004;32:35-42. [PubMed]
- Xie W, Radominska-Pandya A, Shi Y, et al. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci U S A 2001;98:3375-80. [PubMed]
- Xie W, Barwick JL, Simon CM, et al. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev 2000;14:3014-23. [PubMed]
- Choi M, Yamamoto K, Itoh T, et al. Interaction between vitamin D receptor and vitamin D ligands: two-dimensional alanine scanning mutational analysis. Chem Biol 2003;10:261-70. [PubMed]
- Adachi R, Shulman AI, Yamamoto K, et al. Structural determinants for vitamin D receptor response to endocrine and xenobiotic signals. Mol Endocrinol 2004;18:43-52. [PubMed]
- Adachi R, Honma Y, Masuno H, et al. Selective activation of vitamin D receptor by lithocholic acid acetate, a bile acid derivative. J Lipid Res 2005;46:46-57. [PubMed]
- Jurutka PW, Thompson PD, Whitfield GK, et al. Molecular and functional comparison of 1,25-dihydroxyvitamin D(3) and the novel vitamin D receptor ligand, lithocholic acid, in activating transcription of cytochrome P450 3A4. J Cell Biochem 2005;94:917-43. [PubMed]
- Williams ET, Leyk M, Wrighton SA, et al. Estrogen regulation of the cytochrome P450 3A subfamily in humans. J Pharmacol Exp Ther 2004;311:728-35. [PubMed]
- He P, Court MH, Greenblatt DJ, et al. Factors influencing midazolam hydroxylation activity in human liver microsomes. Drug Metab Dispos 2006;34:1198-207. [PubMed]
- Shimada T, Yamazaki H, Mimura M, et al. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 1994;270:414-23. [PubMed]
- Relling MV, Nemec J, Schuetz EG, et al. O-demethylation of epipodophyllotoxins is catalyzed by human cytochrome P450 3A4. Mol Pharmacol 1994;45:352-8. [PubMed]
- Watkins PB, Murray SA, Winkelman LG, et al. Erythromycin breath test as an assay of glucocorticoid-inducible liver cytochromes P-450. Studies in rats and patients. J Clin Invest 1989;83:688-97. [PubMed]
- Kolars JC, Schmiedlin-Ren P, Schuetz JD, et al. Identification of rifampin-inducible P450IIIA4 (CYP3A4) in human small bowel enterocytes. J Clin Invest 1992;90:1871-8. [PubMed]
- Flockhart DA, Rae JM. Cytochrome P450 3A pharmacogenetics: the road that needs traveled. Pharmacogenomics J 2003;3:3-5. [PubMed]
- Mizutani T. PM frequencies of major CYPs in Asians and Caucasians. Drug Metab Rev 2003;35:99-106. [PubMed]
- González R, Cruz A, Ferrín G, et al. Cytoprotective properties of rifampicin are related to the regulation of detoxification system and bile acid transporter expression during hepatocellular injury induced by hydrophobic bile acids. J Hepatobiliary Pancreat Sci 2011;18:740-50. [PubMed]
- Zilly W, Breimer DD, Richter E. Stimulation of drug metabolism by rifampicin in patients with cirrhosis or cholestasis measured by increased hexobarbital and tolbutamide clearance. Eur J Clin Pharmacol 1977;11:287-93. [PubMed]
- Castagnola E, Viscoli C. Rifampicin as treatment for pruritus in malignant cholestasis: an example of an improper indication for anti-infective drugs. Support Care Cancer 1999;7:160. [PubMed]
- Price TJ, Patterson WK, Olver IN. Rifampicin as treatment for pruritus in malignant cholestasis. Support Care Cancer 1998;6:533-5. [PubMed]
- Marschall HU, Wagner M, Zollner G, et al. Complementary stimulation of hepatobiliary transport and detoxification systems by rifampicin and ursodeoxycholic acid in humans. Gastroenterology 2005;129:476-85. [PubMed]
- Wietholtz H, Marschall HU, Sjövall J, et al. Stimulation of bile acid 6 alpha-hydroxylation by rifampin. J Hepatol 1996;24:713-8. [PubMed]
- Galetin A, Ito K, Hallifax D, et al. CYP3A4 substrate selection and substitution in the prediction of potential drug-drug interactions. J Pharmacol Exp Ther 2005;314:180-90. [PubMed]


