
Impact of circulating tumor DNA early detection and serial monitoring in the management of stage I to III colorectal cancer
The temporal course and severity of metastasis to distant organs is a complex process depending on multiple features varying in different cancers and individuals. In patients previously treated for a localized tumor, the latency period before clinical evidence of metastatic relapse is made up of a sequence of events with unpredictable temporal kinetics (1). Until recently, there was no biological or radiological tool that can accurately detect the presence of disseminated tumor cells in distant organs after the primary tumor treatment. The detection of these cells, which constitutes the “minimal residual disease” (MRD—a concept primarily developed in Hematology) is of critical interest, as it would allow tailoring both adjuvant medical therapies and patient follow-up. In that context, circulating tumor biomarkers (a term that encompasses any tumor-derived biomarker that can be detected in a patient’s blood) have been investigated as a possible tool to detect the MRD. Firstly, serum tumor antigens used in the routine follow-up somehow failed to reach theses aims, due to their low specificity and the limited lead time between their rise and relapse detection. In the specific setting of stage I to III colorectal cancer (CRC), post-operative carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) revealed low sensitivity, specificity, positive and negative predictive values to predict metastatic relapses (2). More recently, technological developments allowed to assess the value of other circulating tumor biomarkers, such as circulating tumor cells (CTCs), circulating tumor DNA (ctDNA) and tumor-derived exosomes (3). While tumor-derived exosomes remain very challenging to quantify (4), CTC detection was found to be a very rare event in non-metastatic cancers (5,6). ctDNA, which corresponds to fragmented DNA shed in the blood by dying tumor cells, was found in higher amount than CTC (7-9) and has therefore become the most promising biomarker for MRD detection.
In a prospective, multicenter cohort study on 130 patients that all underwent surgery for a stage I-III CRC, Reinert et al. found that ctDNA detection in the post-operative setting (first month after surgery) and after adjuvant chemotherapy (ACT) was an excellent marker of later relapse: 14 of 16 relapses (87.5%) were identified (10). Patients tested ctDNA-positive in the first month after surgery (and before ACT) were 7 times more likely to relapse than ctDNA-negative patients [hazard ratio (HR) =7.2, 95% CI: 2.7–19.0]. After ACT, ctDNA-positive patients were 17 times (HR =17.5, 95% CI: 5.4–56.5) more likely to relapse. During surveillance, ctDNA-positive patients were more than 40 times more likely to experience disease recurrence than ctDNA-negative patients (HR =43.5; 95% CI: 9.8–193.5). Longitudinal ctDNA analyses revealed disease recurrence up to 16.5 months ahead of standard-of-care radiologic imaging (mean =8.7 months; range, 0.8–16.5 months).
This study therefore confirmed the significant association between ctDNA detection and recurrence-free survival after surgery for CRC. It also highlighted the validity of serial ctDNA testing to monitor CRC relapse. These results are in accordance with previous studies exploring the clinical validity of ctDNA detection in non-metastatic CRC (11-17), but also in breast, lung, or ovarian locally advanced cancer (18). However, the serial ctDNA analysis missed 2 metastatic relapses, despite the fact that the whole-exome sequencing, which is one of the most appropriated genome-wide untargeted approaches, confirmed the presence of the mutations used for plasma screening. This could be explained by the fact that, even in patients with overt metastatic CRC, ctDNA levels may be below the detection limit of our current technological approaches (19,20).
The above-presented results raised the question of the clinical utility of such ctDNA-based monitoring, that is how its actual use could be associated to patient outcome efficacy. Several randomized trials are currently exploring the clinical impact of serial ctDNA analysis in CRC (Table 1).

Full table
In locally advanced CRC, one crucial question is the potential benefit in overall survival of the precociously detection of micrometastatic disease when it leads to more intensive therapeutic strategies. A phase III clinical trial sponsored by the Massachusetts General Hospital (NCT 03803553) is currently evaluating the impact of additional therapy in patients with stage III CRC and a positive ctDNA-test after the completion of standard ACT (Figure 1). Even if this category of patients is systematically treated with ACT according to the current guidelines, the DYNAMIC III trial plans to refine the population of stage III CRC patients who could truly benefit from chemotherapy by comparing ACT guided by ctDNA detection to ACT based on clinical criteria.
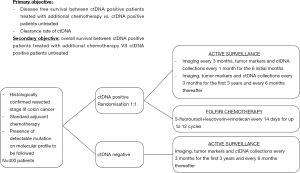
The need for better predictive approaches is deeply required to optimize the selection of patients who could benefit from ACT in stage II CRC. In this population, although the multiple clinicopathological markers validated to distinguish low- and high-risk groups, up to 40% of patients undergo ACT, with significant long-term side effects, for an absolute risk reduction of only 3% to 5%. The IMPROVE-IT trial (NCT 03748680) aims to redefine the population who can really benefit from ACT by comparing ACT plus intensified follow-up to intensified follow-up alone in stage I to II CRC patients, with detectable ctDNA two weeks after surgery and no indication to ACT according to the DCCG (Dutch Colorectal Cancer Group) and ESMO guidelines. The DYNAMIC-rectal trial (ACTRN12617001560381) is an ongoing Australian trial exploring the impact on disease free survival in stage II colon cancer patients randomly assigned to usual care versus ctDNA-informed adjuvant therapy. The NRG Oncology phase II/III trial (COBRA), applying the same design will enroll US and Canadian patients. Positive results could lead future recommendations to include tailored surveillance strategies based on individualized assessment of recurrence risk. The management of serial ctDNA in routine practice could raise questions about the adequate schedule for blood samples collection during patients’ surveillance. The methodology proposed by Reinert et al is clinically relevant, given its concordance to the usual rhythm of post operative follow-up and the short half-life of approximately 2 hours of ctDNA that guarantee its quality of real-time marker. Such a follow-up may remain independent of the initial TNM stage since further work is still needed to determine how well the level of ctDNA correlates with tumor burden. Indeed, Bettegowda et al. emphasized the fact that even if the fraction of patients with detectable ctDNA increased with stage, the level of ctDNA detection also vary within each stage (9).
In conclusion, ctDNA detection commonly untitled “liquid biopsy” represents a convenient and highly repeatable method to overcome issues due to the need to describe the spatial and temporal genomic evolution of tumors from the earliest stage of the neoplastic process to the metastatic disease. Beyond the accuracy of this diagnostic tool, remains the question of its clinical potential applications. The upcoming results of the current controlled prospective clinical trials assessing the utility of ctDNA should give objective answers about the real benefit of ctDNA in CRC and about its place in the clinical decision process.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 2009;9:274-84. [Crossref] [PubMed]
- Moertel CG, Fleming TR, Macdonald JS, et al. An evaluation of the carcinoembryonic antigen (CEA) test for monitoring patients with resected colon cancer. JAMA 1993;270:943-7. [Crossref] [PubMed]
- Speicher MR, Pantel K. Tumor signatures in the blood. Nat Biotechnol 2014;32:441-3. [Crossref] [PubMed]
- Whiteside TL. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv Clin Chem 2016;74:103-41. [Crossref] [PubMed]
- Thery L, Meddis A, Cabel L, et al. Circulating Tumor Cells in Early Breast Cancer. JNCI Cancer Spectr 2019;3: pkz026.
- Cabel L, Proudhon C, Mariani P, et al. Circulating tumor cells and circulating tumor DNA: What surgical oncologists need to know? Eur J Surg Oncol 2017;43:949-62. [Crossref] [PubMed]
- Dawson SJ, Tsui DWY, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209. [Crossref] [PubMed]
- Madic J, Kiialainen A, Bidard F-C, et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer 2015;136:2158-65. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Lin J-K, Lin P-C, Lin C-H, et al. Clinical relevance of alterations in quantity and quality of plasma DNA in colorectal cancer patients: based on the mutation spectra detected in primary tumors. Ann Surg Oncol 2014;21 Suppl 4:S680-6. [Crossref] [PubMed]
- Lecomte T, Berger A, Zinzindohoué F, et al. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer 2002;100:542-8. [Crossref] [PubMed]
- Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. [Crossref] [PubMed]
- Schøler LV, Reinert T, Ørntoft M-BW, et al. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin Cancer Res 2017;23:5437-45. [Crossref] [PubMed]
- Reinert T, Schøler LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016;65:625-34. [Crossref] [PubMed]
- Bazan V, Bruno L, Augello C, et al. Molecular detection of TP53, Ki-Ras and p16INK4A promoter methylation in plasma of patients with colorectal cancer and its association with prognosis. Results of a 3-year GOIM (Gruppo Oncologico dell’Italia Meridionale) prospective study. Ann Oncol 2006;17 Suppl 7:vii84-90. [Crossref] [PubMed]
- Frattini M, Balestra D, Verderio P, et al. Reproducibility of a semiquantitative measurement of circulating DNA in plasma from neoplastic patients. J Clin Oncol 2005;23:3163-4; author reply 3164-5. [Crossref] [PubMed]
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017. [Crossref] [PubMed]
- Bidard FC, Kiavue N, Ychou M, et al. Circulating Tumor Cells and Circulating Tumor DNA Detection in Potentially Resectable Metastatic Colorectal Cancer: A Prospective Ancillary Study to the Unicancer Prodige-14 Trial. Cells 2019. [Crossref] [PubMed]
- Bachet JB, Bouché O, Taieb J, et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: the AGEO RASANC prospective multicenter study. Ann Oncol 2018;29:1211-9. [Crossref] [PubMed]


