
Metabolic biomarker signature for predicting the effect of neoadjuvant chemotherapy of breast cancer
Introduction
Neoadjuvant chemotherapy (NCT) of breast cancer has been used not only for tumor stage, reducing to enable operations but also for improving survival outcomes. Breast cancer patients who could get the pathological complete response (PCR) after NCT were more likely to have improved survival outcomes, whatever the subgroups of HR-positive/HER2-negative, HER2-positive, and triple-negative were, according to a meta-analysis recorded on the 2018 San Antonio Breast Cancer Symposium (SABCS). Similarly, a previous trial presented that all molecular subtypes could gain the same PCR, proved that PCR is an independent predictor of better survival outcomes (1). Thus, PCR has been widely admitted as a surrogate endpoint for predicting the treatment effects on survivals. Nevertheless, just a limited number of patients can achieve PCR, accounting for 30% only (2-4), especially in HER2-negative and hormone-receptor-positive tumors (2). Therefore, beyond consideration of intrinsic tumor subtypes, we may look for other methods to predict response to NCT, thus develop personalized treatment and improve survival rates.
Clinically, researchers tend to evaluate tumor size when patients are accepting NCT and predict the response and PCR possibility. A systematic review shows that magnetic resonance imaging (MRI) is better than the other physical examination, mammography, and ultrasound (US) to evaluate residual disease after NCT (5). Kim et al. classified the tumor shrinkage pattern after NCT of breast cancer patients into four types, however, not all of which changed associating with the difference between pathological responders and non-responders. The overestimation and underestimation of MRI for residual tumor size after NCT always exist, as different histopathological changes would happen in tumors with their different intrinsic types, and treated by different regimens (6). To observe an exhibition or reduction of the serum levels of MUC-1 antigen [cancer antigen (CA) 15.3] before and after NCT is also used to predict the response of NCT (7,8), however, CA 15.3 often expresses in a normal range both before and after treatment in practice. Also, these studies about imaging tools, laboratory indexes can only be employed after a chemotherapy regimen have carried out.
Metabolomics (or metabolite profiling), is a new omics used for biomarker discovery in bio-fluids and tissues, which used more and more widely for detecting disease and predicting drug response and toxicity (9,10). It complements transcriptomics and proteomics to investigate the mechanisms of treatment outcome by directly reflecting biochemical processes. To discover biomarkers for breast cancer, this omics gives messages about both genetics and the environment, which abut the real world better. Additionally, profiles or biomarkers generated from metabolomics tests are cheap and can be more quickly obtained and automated (11).
Metabolomics can be employed to diagnose breast cancer, including the early discovery and the classification of metastatic and early breast cancer, etc. (12,13). However, few studies focus on using metabolomics to predict the effect of chemotherapy of breast cancer. Wei etc. developed a model to predict the chemotherapy of breast cancer by combining the metabolites derived from nuclear magnetic resonance (NMR) and mass spectrometry (MS), achieved a sensitivity of 80%, that is: the model can screen out 80% of the patients who won’t get a PCR in the NCT (14). These results are the beginning and show promise that metabolomics can be applied for assessing the effect of treatment.
This study aimed to use liquid chromatography-mass spectrometry (LC-MS) to investigate the effect of neoadjuvant therapy on metabolic profiles, proving the metabolic differences between responders and non-responders. Serum samples of 35 patients obtained before and after NCT respectively have been studied using LC-MS and then proceeded by multivariate statistics methods. Nine metabolites identified from LC-MS methods divided partial response (PR) and stable disease (SD) patients well. They make out a statistical model, and that with high sensitivity and specificity, to predict the effect of neoadjuvant therapy.
Methods
Sample preparation
From 2014 to 2015, the patients treated at the Department of Mammary Disease, Guangdong Provincial Hospital of Chinese Traditional Medicine, were enrolled in this study. The chemotherapy regimens were decided following the guidelines for neoadjuvant therapy (NCCN.2013.v2). The inclusion criteria included: (I) eligible for NCT (locally advanced breast cancer or patients who hope for a breast-conserving surgery); (II) signed informed consent in person; (III) at least 18 years old and less than 70 years old; (IV) not pregnant before enrolled in the study.
Case report forms (CRF) were used to record the patient’s study data. All patients owned an identification log maintained by the researchers. Assessment measures for the clinical response of NCT in breast cancer patients included three-dimensional (3D) US and MRI. According to resist 1.1, there’s three results of chemotherapy responses came out: PCR, PR (tumor volume reduces exceeding 30%) or SD (tumor volume increases less than 20% or reduces less than 30%). Here, PCR needs no residual cancer both in the breast and axillary lymph nodes, including invasive cancer and carcinoma in situ.
Serum samples of 35 patients of breast cancer were preserved. They were obtained 1 hour before NCT start and 10 days after the final cycle of NCT, respectively. Of these patients, 0 patients enter the PCR group, 16 into the SD group and 19 into PR group. The baseline characteristic of all the eligible patients are displayed below (Table 1).
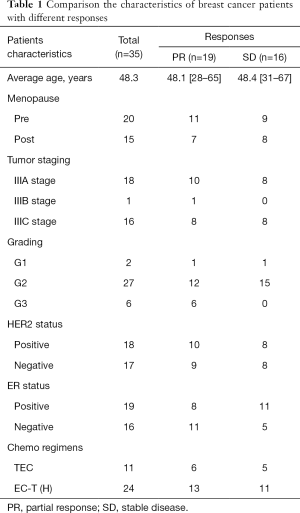
Full table
The collected blood was delivered to the laboratory at once for centrifugation (speed: 10 minutes at 3,000 rpm; temperature: room temperature). Then we got the upper serum constituent, separated and frozen at –80 °C until the final test.
The chemotherapy regimens were drawn up according to the neoadjuvant/adjuvant therapy regimens of NCCN guidelines breast cancer version 2.2013 including (regimen I): EC-T (doxorubicin 90 mg/m2 plus cyclophosphamide 600 mg/m2 IV day 1 every 21 days for 4 cycles followed by docetaxel 100 mg/m2 IV day 1 every 21 days for 4 cycles); (regimen II): TEC (docetaxel plus doxorubicin with the same dose of 75 mg/m2 plus cyclophosphamide 600 mg/m2 every 21 days for 6 cycles). Trastuzumab would employ in HER2/neu positive patients (the dose was 8 mg/kg for the first use; the followed dose was 6 mg/kg, i.e., every 21 days for 1 year). Trastuzumab was not used with anthracyclines simultaneously.
The previously saved serum samples were thawed at 4 °C, 200 µL of it was taken out and placed in 1.5 mL EP tube, mixed with 800 µL of acetonitrile, vortexed for 2 minutes, and then centrifuged at 13,000 rpm for 20 mins at 4 °C. After centrifugation, the supernatant was collected in injection vial for LC-MS analysis.
LC-MS methods
C18 column (100×2.1 mm, 1.7 µm) made by Waters corporation was used for chromatographic separation. Column temperature 30 °C and flow rate was 400 µL/min, sample injection volume 5 µL, sample room temperature 4 °C. The mobile phase: a phase was 0.1% formic acid aqueous, and B phase was 0.1% formic acid acetonitrile separately, gradient elution was used for the sample, and the gradient was set as Table 2 shown. Positive ion mode was used for MS, and the parameters were listed as below, shown in Table 3. The typical metabolism profile chromatograms operating under positive ion mode were illustrated in Figure 1. They respectively represent the typical metabolism profile chromatograms from serum samples of SD patients before chemotherapy (Figure 1A), serum samples of SD patients after chemotherapy (Figure 1B), serum samples of PR patients before chemotherapy (Figure 1C), and serum samples of PR patients after NCT (Figure 1D).
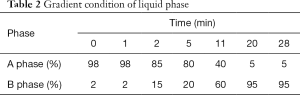
Full table
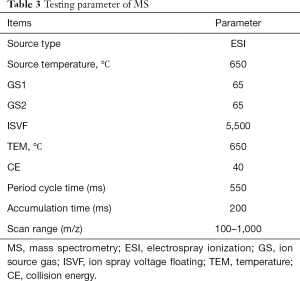
Full table
Data pre-processing
The raw data were gathered by Analyst Software (version 1.5.1, AB SCIEX), made into metabolism profile chromatograms, which were processed by the MarkerViewTM software (version 1.2.1, AB SCIEX). Chromatography combination, the peaks analysis and normalizations of raw data were evaluated in MarkerViewTM Software. Specific settings were as follows: the scope of data collection for 1–28 min, minimum peak strength set on 10% of the base, the minimum peak width set at 5 ppm, the retention time window set at 0.5 min, m/z deviation set at 10 ppm.
Statistical analysis
Least squares discriminant analysis (DA)
The pre-processed quantitative-information data of each sample was imported into SIMCA-P14.0 software for multidimensional statistics, obtaining spectral scores, which were analyzed by partial least squares discriminant analysis (PLS-DA) technique. Then the differences of serum metabolic components between two groups with different outcomes were compared to obtain a set of peaks that contributed the most different to the group. Combining with t-test and other statistics, we obtained the clustering between samples under different grouping conditions, and find out the small molecular compounds in vivo related to the grouping.
According to the contribution degree of the compounds in the sample clustering, the compounds obtained by pattern recognition analysis were analyzed between each group. For the different aspect of clinical significance, we would find the compounds that contribute more to different group of chemotherapy response. Finally, the website HMDB was employed for identifying metabolites. It connects an exact mass spectrum to a special compound or a group of isomerides. Student’s t-test was applied to compare PR and SD groups of samples, and metabolites with significant difference (P<0.01) between two groups were sustained as biomarker candidates for later statistical analysis. They would finally be devoted to building a receiver operating characteristics (ROC) by the SPSS 18.0.
Results
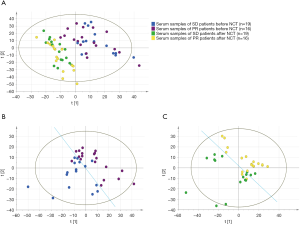
PLS-DA was used to perform the differences of signatures between two groups visually. Results show that chemotherapy could undoubtedly impact the metabolic activities that the two groups both had major change after NCT (Figure 2A). And it is surprising that differences had occurred before NCT start, signatures of PR and SD groups were classified obviously before NCT (Figure 2B); also, they are separated after NCT (Figure 2C). Nevertheless, as we aimed to predict the response before NCT start, we emphasized the result of Figure 2B, which shows the difference between PR and SD groups before NCT start. It illuminated a mechanism that metabolic activities in vivo would result in indifferent response.
According to the values of variable importance in projection (VIP) in the PLS-DA model (VIP >1) and t-test results, we screened out 28 compounds of significant difference (P<0.05) between the two groups. Then an elastic net algorithm was used to construct an effective diagnostic model to predict NCT response. Based on the model, nine serum metabolites were selected. The metabolites identified were mostly lipids, fatty acids, and amino acids, as nutrients of the human body, or to take part in the activities of cell signal transduction, immune regulation, and so on (shown in Table 4).

Full table
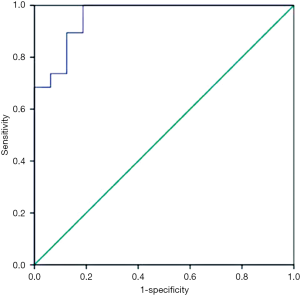
To convert the nine metabolites into a tool for predicting NCT response, a logistics analysis was applied to construct a model by making the ROC curve. With an area under the curve (AUC) of 0.957, The model has a specificity of 100% and sensitivity of 81.2% for predicting the response of PR and SD of breast cancer patients (Figure 3), well proving that these metabolites would be the Metabolic biomarker signatures for predicting response before it took up.
Discussion
Our study was designed to select the potential breast cancer patients who are the potential one to get a PCR after NCT, since it may exclude out those patients who have drug resistance and are originally able to accept a complete resection at the diagnosed time, thus reducing adverse events. The two-two comparisons between SD and PR group before and after NCT performed that metabolic activities of the two groups’ patients before chemotherapy had already distinguishing that would result in a different response. If we find out the metabolic biomarker contributed to the difference of the two groups, we will illustrate some mechanisms why patients have different effects after NCT, even they are in the same subtype, or accept the same chemo-regimens. By LC-MS analytical platform and PLS-DA model, we finally identify nine significant metabolic compounds and make the ROC curve, of which AUC was 0.957, obviously higher than that of Wei’s study, which offered 4 metabolic makers and get an AUC of 0.81 (14). Our model could correctly separate PR from SD patients with higher specificity (100%) and sensitivity (81.2%) (Figure 3). Such a good predictability would undoubtedly further verify a fact that the serum-derived metabolites could be used to predict the effect of breast cancer NCT, ensuring more personalized regimens for patients.
Nine metabolites identified in this study, prostaglandin C1, ricinoleic acid, oleic acid amide, ethyl docosahexaenoic, hulupapeptide, lysophosphatidylethanolamine 0:0/22:4, cysteinyl-lysine, methacholine, and vitamin K2 made up to the prediction model NCT outcome. However, so far, we still do not know how these specific metabolites work to predict the response of chemotherapy.
Of all the metabolites, some have been proved to associate with cancer. Prostaglandin C1 was one of proteinoids. Studies showed that proteinoids would concentrate on patients diagnosed with cancers (15). Prostaglandin E2 (PGE2), another typical proteinoid, has been verified to induce tumor cell invasion in the EP4-mediated pathway, which would account for a worse clinical outcome (16). Another study demonstrated that PGE2 tend to accumulate in the highly metastatic C3L5 and MDA-MB-231 cells, and also, EP4, one of the subtypes of cell surface receptors, have a great impact on the PGE2-induced migration (17). Li’s study reveals that oleic acid is one of the important material to keep malignancy for metastatic carcinoma cells of breast cancer and gastric cancer in an AMPK-dependent pattern. Thus, fatty acid oxidation makes important contribution to cancer cell function (18).
Recently, more and more studies have found that vitamin K2 can result in growth inhibition of a variety of malignant tumor cell growth inhibition, including colon cancer, lung cancer, breast cancer cells (19,20), and induce the apoptosis of liver cell cancer, ovarian cancer, and another solid tumor, leukemia, myelodysplastic syndrome. Its synergy with a variety of antitumor drugs. Vitamin K2 and its derivatives have the inhibitory effect on the liver cancer cells, influence the postoperative recurrence and overall survival of liver cancer (21). However, the mechanism about vitamin K2 antitumor is still disputed, it may act by involving in mitochondrial electron signal transmission, affecting the Bcl-2 protein, inducing cell apoptosis of caspase family, adjusting the G1 phase-relevant molecular expression of cell cycle, etc. (22,23).
Our study has limitations. The sample size of this study was only 35 patients, and it should be necessary to expand the sample size to verify the above results and special metabolites, such as vitamin K2, be quantified for further study. Moreover, an independent sample cohort is essential to be the validation set. Thus, we can further understand the molecular basis of the different effects between two groups of patients. Notwithstanding, these reassuring results, offer a new approach to decide which cohorts should accept NCT deliberately, beyond their intrinsic subtypes. Reassuringly, we found some metabolites associated closely to the different outcome of breast cancer NCT, making them as hints for further molecular studies.
Conclusions
To predict the effect of NCT of breast cancer is of great significance, as a special effect would associate with a long-term survival of breast cancer. In this article, we present a prediction model for the effect of breast cancer NCT based on LC-MS metabolic profiling methods. Nine metabolites selected made up to build the model, distinguishing groups of patients with partial or steady response. The results demonstrate the metabolomics is promising for the prediction of the treatment response.
Acknowledgments
Funding: This study was supported by the foundation of Guangdong Provincial Administration of Traditional Chinese Medicine, China (grant number: 20123008, 20132159). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. During the period of 2010–2015, our institution successively passed the ethics certification of WHO-SIDCER and Ethics Research System of Traditional Chinese Medicine. After that, our institution mandated that all human studies should be ethically reviewed, that is, the first case collection started on January 1, 2016. The cases collected in this article were from 2014 to 2015, and no ethical approval certificate was obtained at that time.
References
- Bonnefoi H, Litière S, Piccart M, et al. Pathological complete response after neoadjuvant chemotherapy is an independent predictive factor irrespective of simplified breast cancer intrinsic subtypes: a landmark and two-step approach analyses from the EORTC 10994/BIG 1-00 phase III trial. Ann Oncol 2014;25:1128-36. [Crossref] [PubMed]
- Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014;384:164-72. [Crossref] [PubMed]
- Houssami N, Macaskill P, von Minckwitz G, et al. Meta-analysis of the association of breast cancer subtype and pathologic complete response to neoadjuvant chemotherapy. Eur J Cancer 2012;48:3342-54. [Crossref] [PubMed]
- Jones RL, Smith IE. Neoadjuvant treatment for early-stage breast cancer: opportunities to assess tumour response. Lancet Oncol 2006;7:869-74. [Crossref] [PubMed]
- Lobbes MB, Prevos R, Smidt M, et al. The role of magnetic resonance imaging in assessing residual disease and pathologic complete response in breast cancer patients receiving neoadjuvant chemotherapy: a systematic review. Insights Imaging 2013;4:163-75. [Crossref] [PubMed]
- Kim TH, Kang DK, Yim H, et al. Magnetic resonance imaging patterns of tumor regression after neoadjuvant chemotherapy in breast cancer patients: correlation with pathological response grading system based on tumor cellularity. J Comput Assist Tomogr 2012;36:200-6. [Crossref] [PubMed]
- Al-azawi D, Kelly G, Myers E, et al. CA 15-3 is predictive of response and disease recurrence following treatment in locally advanced breast cancer. BMC Cancer 2006;6:220. [Crossref] [PubMed]
- Kurebayashi J, Yamamoto Y, Tanaka K, et al. Significance of serum carcinoembryonic antigen and CA 15-3 in monitoring advanced breast cancer patients treated with systemic therapy: a large-scale retrospective study. Breast Cancer 2003;10:38-44. [Crossref] [PubMed]
- Clayton TA, Lindon JC, Cloarec O, et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006;440:1073-7. [Crossref] [PubMed]
- Gowda GA, Zhang S, Gu H, et al. Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn 2008;8:617-33. [Crossref] [PubMed]
- Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res 2009;15:431-40. [Crossref] [PubMed]
- Oakman C, Tenori L, Claudino WM, et al. Identification of a serum-detectable metabolomic fingerprint potentially correlated with the presence of micrometastatic disease in early breast cancer patients at varying risks of disease relapse by traditional prognostic methods. Ann Oncol 2011;22:1295-301. [Crossref] [PubMed]
- Slupsky CM, Steed H, Wells TH, et al. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin Cancer Res 2010;16:5835-41. [Crossref] [PubMed]
- Wei S, Liu L, Zhang J, et al. Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol Oncol 2013;7:297-307. [Crossref] [PubMed]
- Asano T, Shoda J, Ueda T, et al. Expressions of cyclooxygenase-2 and prostaglandin E-receptors in carcinoma of the gallbladder: crucial role of arachidonate metabolism in tumor growth and progression. Clin Cancer Res 2002;8:1157-67. [PubMed]
- Wu J, Zhang Y, Frilot N, et al. Prostaglandin E2 regulates renal cell carcinoma invasion through the EP4 receptor-Rap GTPase signal transduction pathway. J Biol Chem 2011;286:33954-62. [Crossref] [PubMed]
- Timoshenko AV, Xu G, Chakrabarti S, et al. Role of prostaglandin E2 receptors in migration of murine and human breast cancer cells. Exp Cell Res 2003;289:265-74. [Crossref] [PubMed]
- Li S, Zhou T, Li C, et al. High metastaticgastric and breast cancer cells consume oleic acid in an AMPK dependent manner. PLoS One 2014;9:e97330. [Crossref] [PubMed]
- Amalia H, Sasaki R, Suzuki Y, et al. Vitamin K2-derived compounds induce growth inhibition in radioresistant cancer cells. Kobe J Med Sci 2010;56:E38-49. [PubMed]
- Xia J, Matsuhashi S, Hamajima H, et al. The role of PKC isoforms in the inhibition of NF-κB activation by vitamin K2 in human hepatocellular carcinoma cells. J Nutr Biochem 2012;23:1668-75. [Crossref] [PubMed]
- Ishizuka M, Kubota K, Shimoda M, et al. Effect of menatetrenone, a vitamin k2 analog, on recurrence of hepatocellular carcinoma after surgical resection: a prospective randomized controlled trial. Anticancer Res 2012;32:5415-20. [PubMed]
- Kitagawa J, Hara T, Tsurumi H, et al. Synergistic growth inhibition in HL-60 cells by the combination of acyclic retinoid and vitamin K2. J Cancer Res Clin Oncol 2011;137:779-87. [Crossref] [PubMed]
- Ying WX, Chen H, Li RZ. Effect of Vi-tamin K2 and benazepril combination therapy on gas-tric cancer in nude mice. Chinese Pharmaceutical Journal 2012;47:186-9.


