Genome-wide association study of cerebrospinal fluid neurofilament light levels in non-demented elders
Introduction
Neurofilaments are major structural components of neuronal cytoskeleton, comprising neurofilament heavy, neurofilament medium and neurofilament light (NFL). As an emerging biomarker for axonal damage and neuronal death, NFL is primarily expressed in large-caliber myelinated axons in the subcortical brain regions (1). When axonal membrane integrity is disrupted, NFL will be released into the extracellular space and may diffuse into cerebrospinal fluid (CSF) (2-4). CSF NFL concentrations are associated with varieties of pathological conditions, including Alzheimer’s disease (AD), frontotemporal dementia and amyotrophic lateral sclerosis (2,5-8). Further, elevated CSF NFL levels are also found in normal cognitive aging and early clinical stage of AD (5,7,9). They are associated with brain atrophy and could predict hippocampal atrophy rate (5,10,11). Little is known about the early influence of genetic factors on the CSF NFL levels. Identifying variants that influence CSF NFL levels during the stages preceding dementia may provide insight into the molecular processes underlying the brain aging and etiology of neurodegenerative diseases, and may ultimately help delay or prevent diseases.
Here, we performed a genome-wide association study (GWAS) of non-demented elders to identify novel genetic variants that modulate the levels of CSF NFL. In addition, we further investigated whether the identified genetic variants have association with other AD features.
Methods
Alzheimer’s disease neuroimaging initiative (ADNI)
Data used in this study were obtained from the ADNI database. ADNI is an international longitudinal study with approximately 50 sites which was initiated in 2003 across the United States and Canada. The initial aim of ADNI is to identify and investigate whether the combination of neuroimaging, genetics, other biological markers and neuropsychological tests can measure the progression of mild cognitive impairment (MCI) and early AD (12). ADNI has covered more than 1,500 participants including cognitively normal (CN), MCI and early AD (13). Further information about ADNI can be found online (http://adni-info.org/).
Subjects
Individuals from ADNI were included in our study if they were judged clinically as CN or MCI at baseline. A total of 977 non-demented participants who had available baseline and 7-year follow-up data remained in this study. The number of patients who had data of CSF NFL is limited in the ADNI database. The data of CSF NFL and genotype was available from 285 participants who are non-demented elders. To reduce the potential influence of population stratification, all participants of our genetic analyses were restricted to non-Hispanic white individuals. Multidimensional scaling (MDS) plot was performed to confirm the population substructure (Figure S1). Finally, 263 participants who passed the quality control (QC) were included in GWAS analysis.
Participants of MCI was included if with memory complaint, a Mini-Mental State Exam (MMSE) score between 24 and 30, a global Clinical Dementia Rating (CDR) score of 0.5, a CDR memory score of 0.5 or greater and abnormal memory function on the Wechsler Memory Scale-Logical Memory II test. The National Institute on Aging and the Alzheimer’s Association (NIA-AA) criteria has been proposed to identify non-demented subjects with the risk of AD based on the presence of AD biomarkers (14). The stage of preclinical AD (Pre-AD) was intermediate between the appearance of neuropathological brain lesion and the AD symptoms onset (15). During this stage, amyloid positive may be the first signs to occur and can be established using CSF (15,16). The Pre-AD patients were defined as CN participants who had abnormal amyloid, with no impairment in cognition or subtle cognitive decline (14). Therefore, in order to further deepen the understanding of the early asymptomatic stages of AD, Pre-AD was separated from CN as a group for analysis besides the existing groups based on diagnoses. Amyloid positive (A+) and negative (A−) were divided by the proposed CSF amyloid-β (Aβ) cutoff value of 192 pg/mL (17).
Phenotypic evaluation
AD-related phenotypes including CSF biomarkers, cognitive assessment and brain magnetic resonance imaging (MRI) scan data were selected for analysis, all of which were obtained from the ADNI dataset at baseline and during follow-up. CSF samples were collected from ADNI subjects in the morning after an overnight fast. All the CSF samples were collected at University of Pennsylvania Alzheimer’s Disease Clinical Core using standardized methodology including storage of aliquots in polypropylene vials maintained in the repository at −80 °C. CSF concentrations of total tau (t-tau), phosphorylated tau (p-tau) and Aβ were measured using the multiplex xMAP Luminex platform (Luminex Corp, Austin, TX, USA) with the INNO-BIA AlzBio3 kit (Fujirebio, Ghent, Belgium), as described by the manufacturer. CSF NFL levels were determined by a commercial enzyme-linked immunosorbent assay (NF-light ELISA, Uman Diagnostics) (5). The scores of the two cognitive assessments—MMSE and CDR-Sum of Boxes (CDR-SB) were used for the association analysis. And three regional volumes on MRI were analyzed: ventricular, hippocampal volumes and white matter hyperintensities (WMHs). Data on brain MRI measures were obtained with 1.5T or 3.0T MRI scanners. FreeSurfer, version 4.3, was used for quantification of brain regional volumes. WMH volumes were not normally distributed and therefore were transformed using the logarithm. A detailed description of collecting data on CSF biomarkers and brain MRI scan can be found elsewhere (17,18).
Genotyping and QC
Genotyping of the ADNI samples was performed using the Illumina 610-Quad BeadChip (Illumina, Inc., San Diego, CA, USA) (19). The APOE gene has three alleles, APOE ε2 (cys112 and cys158), APOE ε3 (cys112 and arg158) and APOE ε4 (arg112 and arg158), defining six genotypes: ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4 and ε4/ε4. TaqMan Genotyping Technology was used to genotype rs7412 and rs429358, which can determine APOE ε2/ε3/ε4 status. Genotype data underwent stringent QC procedures including: minimum call rate for single nucleotide polymorphisms (SNPs) and individuals >97%, Hardy-Weinberg equilibrium test P>1×10−3, and minor allele frequency (MAF) >0.02. After QC, 441,611 single SNPs were available for analysis.
Statistical analyses
Baseline age, gender, education years, APOE ε4 status and two MDS components were included as covariates in GWAS. Demographic differences between groups were tested using Wilcoxon rank-sum test or Chi-square tests for continuous or categorical variables, respectively. We considered values of P<0.05 as significant and all of the genetic analysis was performed under an additive genetic model. Analysis of associations between SNPs and the phenotype at baseline was carried out using multiple linear regression. Longitudinal analysis of follow-up data was performed using mixed effects models. Baseline age, gender, education years and APOE ε4 status were included as covariates both in cross-sectional and longitudinal analyses. To facilitate the comparisons between modalities, all outcome variables in genetic analysis were standardized to z scores. The Kaplan-Meier survival curves were plotted to estimate the effect of genotype on the risks of progression from no cognitive impairment to the prodromal stage of AD and from MCI to incident AD dementia. And the CDR was used to describe the severity of dementia. Individuals with CDR =0 were categorized as controls, and those with CDR scores higher than 0.5 were defined as the prodromal stage of AD (20). Cox proportional hazards regression was used to investigate which factors were associated with the progression. GWAS and all further analyses were conducted using PLINK and R (Version 3.4.4; The R Foundation). The Manhattan-type and quantile-quantile (Q-Q) plots were generated by a software program (R Studio, version 1.1.383) and regional association plots were visualized with the LocusZoom web tool (http://locuszoom.org/).
Results
Baseline characteristics of included subjects
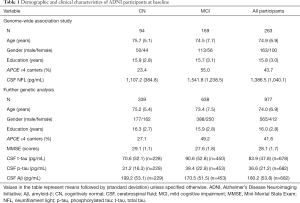
Table 1 summarizes group-wise CSF biomarker variables as well as demographic and psychometric data in the baseline clinical assessment of participants included in the present study. In the samples for GWAS, APOE ε4 status are significantly different (P<0.05) among baseline diagnostic groups (CN and MCI). As for the samples included in further genetic analyses, MCI subgroup were younger (P=0.002) and less educated (P=0.043) with higher frequency of APOE ε4 allele (P<0.001) and worse performance on MMSE (P<0.001). As expected, baseline CSF concentrations of p-tau, t-tau and NFL were higher and Aβ was lower in MCI than in CN (P<0.001 for all).
Full table
GWAS
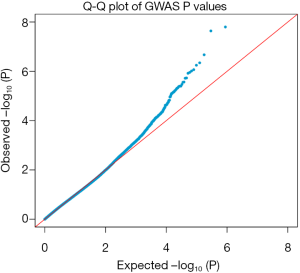
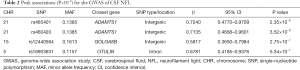
Through GWAS analysis, we identified genome-wide significant association with two novel SNPs (rs465401, rs460420) for CSF NFL at a threshold of P=5×10−8 (Figure 1A and Table 2). The QQ plot is shown in Figure S2. Since the 2 SNPs on chromosome 21 were in high linkage disequilibrium (LD) (r2 =1, D’ =1), we selected one of them with the peak P value (rs465401) for further analysis. The strongest association for CSF NFL is found with rs465401 (P=2.35×10−8), located near the ADAMTS1 (a disintegrin-like and metalloproteinase with thrombospondin type 1 motif) gene. The LD pattern between rs465401 and nearby SNPs is shown in Figure 1B. These nearby SNPs showed association with CSF NFL levels with the P<0.01. After controlling for the top SNP (rs465401), these SNPs associated with CSF NFL also disappeared, indicating that all the associations of this locus were driven by the top SNP (Figure 1C).
Full table

Rs465401 and CSF NFL levels
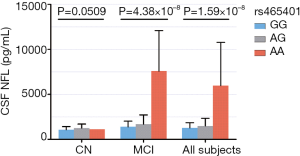
Rs465401 was analyzed further to evaluate possible associations of CSF NFL levels with baseline diagnosis and genotypes (Figure 2). An association was revealed between the minor allele of rs465401 (A) and increased CSF NFL concentration in a dose-dependent effect in a hybrid population without dementia (AA: 5,960.25±4,802.78 pg/mL, AG: 1,480.30±855.34 pg/mL, GG: 1,275.49±52.85 pg/mL) as well as in MCI group (AA: 7,578.33±4,503.64 pg/mL, AG: 1,674.25±1,039.42 pg/mL, GG: 1,383.66±630.40 pg/mL). However, no significant association has been found between rs465401 and CSF NFL levels in CN.
Rs465401 and AD-related phenotypes
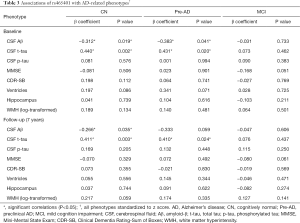
Since the most significant variant associated with CSF levels of NFL was rs465401, we examined the relation of this SNP to other CSF analytes, cognition and neuroimaging measures data (Table 3).
Full table
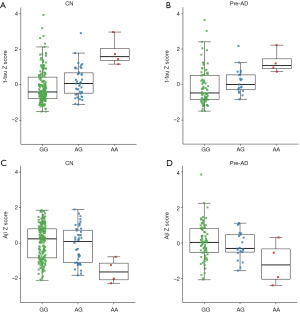
In our primary analysis, we evaluated the associations between the minor allele of rs465401 (A) and AD endophenotypes at baseline (Figure 3). Analysis stratifying the participants based on diagnostic groups showed that rs465401 was associated with CSF t-tau (P=0.002, β=0.440) and Aβ (P=0.019, β=−0.312) in CN group. When taking amyloidosis into account, mutations of rs465401 still had correlations with CSF t-tau (P=0.020, β=0.431) and Aβ (P=0.041, β=−0.383) in Pre-AD group. No associations were observed of rs465401 with MRI scans and cognitive assessment. Although a significant correlation has been reported between CSF NFL concentration and white matter change (5), there was no significant association of rs465401 with log transformed WMH in this study.
In the secondary analysis, longitudinal analysis was performed to evaluated whether rs465401 also influenced these analytes over the follow-up period. The analysis showed that rs465401 was associated with the longitudinal elevation of CSF t-tau (P=0.003, β=0.411) as well as the decrease of CSF Aβ (P=0.035, β=−0.266) in CN. Furthermore, mutations of rs465401 had correlation with an elevation of t-tau (P=0.024, β=0.410) within Pre-AD subgroup.
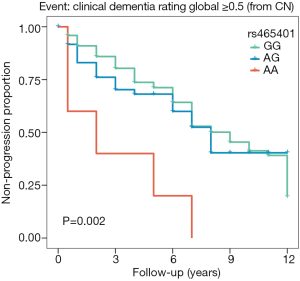
In the tertiary analysis, survival analysis showed that rs465401 was associated with the risk of incident prodromal AD within the follow-up in CN (Figure 4). AA homozygotes had a 3.59 (95% CI: 1.40–9.20) increased hazard, indicating a 3.59 larger relative risk for progression to the prodromal stage of AD compared with GG homozygotes. No difference in the risk of progression from MCI to AD was identified among the three rs465401 carrier statuses.
Discussion
In the present study, we identified two genetic variants (rs465401 and rs460420) near the ADAMTS1 gene that are genome-wide significantly associated with CSF NFL levels in the ADNI samples without dementia. The minor allele (A) of rs465401 was associated with CSF NFL elevation. We also found that greater rs465401 minor-allele (A) dosage was associated with higher CSF Aβ, lower t-tau levels and greater longitudinal change in both Aβ and t-tau within CN participants. CN AA homozygotes had a more than threefold risk of clinical progression within the follow-up compared with GG homozygotes.
The molecular mechanism through which rs465401 could affect CSF levels of NFL has not been studied yet. These SNPs map to an intergenic region near the ADAMTS1 gene on chromosome 21. ADAMTS1 is a protein coding gene which is most known for its effect on atherosclerosis (21), muscle injury (22) and normal follicular development and ovulatory process (23). Only recently more attention is paid to its role in the central nervous system (CNS) (24). The Brain RNA-Seq database (http://web.stanford.edu/group/barres_lab/brainseqMariko/brainseq2.html) shows that ADAMTS1 was primarily expressed in oligodendrocytes and neurons in the temporal cortex (25). This gene encodes a matrix metalloprotease whose natural substrates are proteoglycans (aggrecan and versican) (26,27). Aggrecan and versican are expressed in the brain, belonging to the chondroitin sulphate proteoglycans (CSPGs) family. As a group, CSPGs have been shown to play both neuroprotective and neurotoxic roles within the CNS (28,29). The ADAMTS-mediated CSPG degradation may be favorable to the healing of injured CNS tissue. Meanwhile, it also enables the infiltration of inflammatory cells and potentiate brain injury (30,31). That may lead to neuronal damage and NFL leakage into CSF, suggesting a possible role of ADAMTS1 in CSF NFL elevation. Meanwhile, research also revealed that the area enriched with CSPGs in the extracellular matrix of human cortex rarely undergoes cytoskeletal alterations in AD (28). ADAMTS1, which shows a manifold approximately sevenfold overexpression in AD individuals (32), specifically degrades CSPGs, suggesting a pathological relevance to disease progression of AD. Recently, rs2830500 was identified to be the risk locus of AD, and research nominated ADAMTS1 as a likely risk gene of AD (33). We also explored the associations in ADNI database, however there was no significant association between rs2830500 and CSF NFL levels (P=0.419, β=−0.107).
Combined with previous results, our findings found that rs465401 was significantly associated with CSF levels of t-tau and Aβ among CN and Pre-AD groups, suggesting that ADAMTS1 may play a potential role in AD pathology. T-tau and Aβ are the most well-established AD biomarkers of neurofibrillary tangle pathology and amyloid plaque respectively. T-tau is found at high levels due to cortical neuronal loss in AD patients, and Aβ at low concentrations due to cortical amyloid deposition. ADAMTS1 variants maybe be related to AD via tau pathway and amyloid pathology in the CN stage and influence the disease’s progression prior to extensive and irreversible neural damage.
NFL is released into the extracellular space after axonal damage. The levels of NFL can be measured in both CSF and blood. It has been reported that the levels of NFL in CSF were correlated with those in blood (34). The concentrations of NFL in blood is about 40-fold lower than those in CSF. Li et al. has reported that two variants in LUZP2 gene and GABRB2 gene were associated with plasma NFL at suggestive levels in elders including AD patients (35). But in our study, we have found that rs465401 and rs460420 near ADAMTS1 gene were significantly associated with CSF NFL levels in non-demented elders. It was found that the levels of NFL can be elevated 16.2 years before the estimated symptom onset of AD (36). So, we have restricted the study population to non-demented elders to identify the genetic modifier of NFL in our study. Our findings that variants near ADAMTS1 gene were associated with CSF NFL levels suggested that rs465401 and rs460420 might be regulators of CSF NFL.
More research is needed to elucidate the specific actions of ADAMTS1 in CNS, and understanding the effect of rs465401 on non-demented elders may play a role in risk stratification among non-demented elders. It is also possible that the effect of rs465401 on CSF levels of NFL as well as AD-related biomarkers is through its effect on genes rather than ADAMTS1. Therefore, the exact mechanism needs to be explored in further research.
This study also has some potential limitations. The GWAS sample size was relatively small. That may lead to insufficient power of detecting a statistically significant effect on AD endophenotypes. Thus, replicating these findings with independent, larger samples in future studies will be necessary to help understand the biology underlying pathology. And studies of longitudinal change in NFL levels may reveal additional associations. Another limitation is that the sample was restricted to non-Hispanic white participants to avoid genetic stratification across ethnicities. Further studies are required in other populations.
Conclusions
Overall, our study provides insight into the relationship of genetic variants with CSF NFL and AD-related phenotypes. Although the pathogenic mechanism remains unclear, the associations identified in this study may make the SNPs ideal candidates for future genetic studies on aging and neurodegenerative disorders.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (No. 81771148), Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX01) and ZHANGJIANG LAB, Tianqiao and Chrissy Chen Institute, and the State Key Laboratory of Neurobiology and Frontiers Center for Brain Science of Ministry of Education, Fudan University. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the NIA, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study procedures were approved by the institutional review boards of all participating centers (https://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf), and written informed consent was obtained from all participants or their authorized representatives. Ethics approval was obtained from the institutional review boards of each institution involved: Oregon Health and Science University; University of Southern California; University of California—San Diego; University of Michigan; Mayo Clinic, Rochester; Baylor College of Medicine; Columbia University Medical Center; Washington University, St. Louis; University of Alabama at Birmingham; Mount Sinai School of Medicine; Rush University Medical Center; Wien Center; Johns Hopkins University; New York University; Duke University Medical Center; University of Pennsylvania; University of Kentucky; University of Pittsburgh; University of Rochester Medical Center; University of California, Irvine; University of Texas Southwestern Medical School; Emory University; University of Kansas, Medical Center; University of California, Los Angeles; Mayo Clinic, Jacksonville; Indiana University; Yale University School of Medicine; McGill University, Montreal-Jewish General Hospital; Sunnybrook Health Sciences, Ontario; U.B.C. Clinic for AD & Related Disorders; Cognitive Neurology—St. Joseph’s, Ontario; Cleveland Clinic Lou Ruvo Center for Brain Health; Northwestern University; Premiere Research Inst (Palm Beach Neurology); Georgetown University Medical Center; Brigham and Women’s Hospital; Stanford University; Banner Sun Health Research Institute; Boston University; Howard University; Case Western Reserve University; University of California, Davis—Sacramento; Neurological Care of CNY; Parkwood Hospital; University of Wisconsin; University of California, Irvine—BIC; Banner Alzheimer’s Institute; Dent Neurologic Institute; Ohio State University; Albany Medical College; Hartford Hospital, Olin Neuropsychiatry Research Center; Dartmouth-Hitchcock Medical Center; Wake Forest University Health Sciences; Rhode Island Hospital; Butler Hospital; UC San Francisco; Medical University South Carolina; St. Joseph’s Health Care Nathan Kline Institute; University of Iowa College of Medicine; Cornell University; and University of South Florida: USF Health Byrd Alzheimer’s Institute.
References
- Menke RA, Gray E, Lu CH, et al. CSF neurofilament light chain reflects corticospinal tract degeneration in ALS. Ann Clin Transl Neurol 2015;2:748-55. [Crossref] [PubMed]
- Landqvist Waldo M, Frizell Santillo A, Passant U, et al. Cerebrospinal fluid neurofilament light chain protein levels in subtypes of frontotemporal dementia. BMC Neurol 2013;13:54. [Crossref] [PubMed]
- Norgren N, Rosengren L, Stigbrand T. Elevated neurofilament levels in neurological diseases. Brain Res 2003;987:25-31. [Crossref] [PubMed]
- Zanier ER, Refai D, Zipfel GJ, et al. Neurofilament light chain levels in ventricular cerebrospinal fluid after acute aneurysmal subarachnoid haemorrhage. J Neurol Neurosurg Psychiatry 2011;82:157-9. [Crossref] [PubMed]
- Zetterberg H, Skillback T, Mattsson N, et al. Association of Cerebrospinal Fluid Neurofilament Light Concentration With Alzheimer Disease Progression. JAMA Neurol 2016;73:60-7. [Crossref] [PubMed]
- Pijnenburg YA, Janssen JC, Schoonenboom NS, et al. CSF neurofilaments in frontotemporal dementia compared with early onset Alzheimer's disease and controls. Dement Geriatr Cogn Disord 2007;23:225-30. [Crossref] [PubMed]
- Skillback T, Farahmand B, Bartlett JW, et al. CSF neurofilament light differs in neurodegenerative diseases and predicts severity and survival. Neurology 2014;83:1945-53. [Crossref] [PubMed]
- Zetterberg H, Jacobsson J, Rosengren L, et al. Cerebrospinal fluid neurofilament light levels in amyotrophic lateral sclerosis: impact of SOD1 genotype. Eur J Neurol 2007;14:1329-33. [Crossref] [PubMed]
- Vagberg M, Norgren N, Dring A, et al. Levels and Age Dependency of Neurofilament Light and Glial Fibrillary Acidic Protein in Healthy Individuals and Their Relation to the Brain Parenchymal Fraction. PLoS One 2015;10:e0135886. [Crossref] [PubMed]
- Pereira JB, Westman E, Hansson O. Association between cerebrospinal fluid and plasma neurodegeneration biomarkers with brain atrophy in Alzheimer's disease. Neurobiol Aging 2017;58:14-29. [Crossref] [PubMed]
- Idland AV, Sala-Llonch R, Borza T, et al. CSF neurofilament light levels predict hippocampal atrophy in cognitively healthy older adults. Neurobiol Aging 2017;49:138-44. [Crossref] [PubMed]
- Weiner MW, Aisen PS, Jack CR Jr, et al. The Alzheimer's disease neuroimaging initiative: progress report and future plans. Alzheimers Dement 2010;6:202-11.e7. [Crossref] [PubMed]
- Weiner MW, Veitch DP, Aisen PS, et al. The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement 2013;9:e111-94. [Crossref] [PubMed]
- Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011;7:280-92. [Crossref] [PubMed]
- Dubois B, Hampel H, Feldman HH, et al. Preclinical Alzheimer's disease: Definition, natural history, and diagnostic criteria. Alzheimers Dement 2016;12:292-323. [Crossref] [PubMed]
- Jack CR Jr, Wiste HJ, Weigand SD, et al. Amyloid-first and neurodegeneration-first profiles characterize incident amyloid PET positivity. Neurology 2013;81:1732-40. [Crossref] [PubMed]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol 2009;65:403-13. [Crossref] [PubMed]
- Jack CR Jr, Bernstein MA, Fox NC, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 2008;27:685-91. [Crossref] [PubMed]
- Roostaei T, Nazeri A, Felsky D, et al. Genome-wide interaction study of brain beta-amyloid burden and cognitive impairment in Alzheimer's disease. Mol Psychiatry 2017;22:287-95. [Crossref] [PubMed]
- Donohue MC, Sperling RA, Petersen R, et al. Association Between Elevated Brain Amyloid and Subsequent Cognitive Decline Among Cognitively Normal Persons. JAMA 2017;317:2305-16. [Crossref] [PubMed]
- Jonsson-Rylander AC, Nilsson T, Fritsche-Danielson R, et al. Role of ADAMTS-1 in atherosclerosis: remodeling of carotid artery, immunohistochemistry, and proteolysis of versican. Arterioscler Thromb Vasc Biol 2005;25:180-5. [Crossref] [PubMed]
- Du H, Shih CH, Wosczyna MN, et al. Macrophage-released ADAMTS1 promotes muscle stem cell activation. Nat Commun 2017;8:669. [Crossref] [PubMed]
- Shozu M, Minami N, Yokoyama H, et al. ADAMTS-1 is involved in normal follicular development, ovulatory process and organization of the medullary vascular network in the ovary. J Mol Endocrinol 2005;35:343-55. [Crossref] [PubMed]
- Lemarchant S, Pruvost M, Montaner J, et al. ADAMTS proteoglycanases in the physiological and pathological central nervous system. J Neuroinflammation 2013;10:133. [Crossref] [PubMed]
- Zhang Y, Chen K, Sloan SA, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014;34:11929-47. [Crossref] [PubMed]
- Kuno K, Okada Y, Kawashima H, et al. ADAMTS-1 cleaves a cartilage proteoglycan, aggrecan. FEBS Lett 2000;478:241-5. [Crossref] [PubMed]
- Sandy JD, Westling J, Kenagy RD, et al. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441-Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J Biol Chem 2001;276:13372-8. [Crossref] [PubMed]
- Hartig W, Klein C, Brauer K, et al. Hyperphosphorylated protein tau is restricted to neurons devoid of perineuronal nets in the cortex of aged bison. Neurobiol Aging 2001;22:25-33. [Crossref] [PubMed]
- Jones LL, Margolis RU, Tuszynski MH. The chondroitin sulfate proteoglycans neurocan, brevican, phosphacan, and versican are differentially regulated following spinal cord injury. Exp Neurol 2003;182:399-411. [Crossref] [PubMed]
- Rhodes KE, Fawcett JW. Chondroitin sulphate proteoglycans: preventing plasticity or protecting the CNS? J Anat 2004;204:33-48. [Crossref] [PubMed]
- Cross AK, Haddock G, Stock CJ, et al. ADAMTS-1 and -4 are up-regulated following transient middle cerebral artery occlusion in the rat and their expression is modulated by TNF in cultured astrocytes. Brain Res 2006;1088:19-30. [Crossref] [PubMed]
- Miguel RF, Pollak A, Lubec G. Metalloproteinase ADAMTS-1 but not ADAMTS-5 is manifold overexpressed in neurodegenerative disorders as Down syndrome, Alzheimer's and Pick's disease. Brain Res Mol Brain Res 2005;133:1-5. [Crossref] [PubMed]
- Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet 2019;51:414-30. [Crossref] [PubMed]
- Disanto G, Barro C, Benkert P, et al. Serum Neurofilament light: A biomarker of neuronal damage in multiple sclerosis. Ann Neurol 2017;81:857-70. [Crossref] [PubMed]
- Li JQ, Yuan XZ, Li HY, et al. Genome-wide association study identifies two loci influencing plasma neurofilament light levels. BMC Med Genomics 2018;11:47. [Crossref] [PubMed]
- Preische O, Schultz SA, Apel A, et al. Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer's disease. Nat Med 2019;25:277-83. [Crossref] [PubMed]