
Therapeutic potential of microRNAs in the regulation of cancer energy metabolism
Reprogramming of energy metabolism is one of the hallmarks of cancers caused by genomic instability and contributes to their adaptation within the tumor microenvironment and resistance to anticancer therapy (1). The tricarboxylic acid cycle, aerobic glycolysis, de novo fatty acid synthesis, and altered autophagy allow tumor cells to survive under adverse conditions (2). Epithelial cells primarily store energy as glycogen, a high-density glucose polymer, which provides the organism with an immediate source of glucose to support cellular energy requirements. Epithelial cancer cells reprogram their metabolic pathways to meet these needs during the process of tumor progression. Normal cells consume glucose to generate adenosine triphosphate (ATP) via oxidative phosphorylation under aerobic conditions and switch to glycolysis only under anoxic conditions. However, many tumor cells prefer glycolysis even in the presence of normal oxygen concentration. This phenomenon, known as the Warburg effect, is characterized by increased glycolysis and lactate production regardless of oxygen availability (3), which diverts glucose metabolites from energy production to anabolic processes to accelerate cell proliferation (4). Decades of studies focusing on mitochondrial functions in cancer cells revealed that cells maintain a high rate of oxidative phosphorylation (5). This indicates that the Warburg effect is not a single rule, but mitochondrial functions are also essential for cancer progression (6,7). Another important energy source for cancer growth is glutamine, an amino acid. During malignant transformation, cancer cells modify the consumption and processing of glutamine to sustain cell growth and proliferation (8). It is not only consumed as an energy source but used for the production of lipids, amino acids, and nucleotides for cell replication and the maintenance of mitochondrial membrane potential (9). The outline of energy metabolism in cancer cells is summarized in Figure 1.
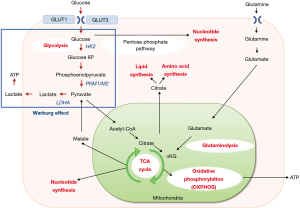
Since the discovery of microRNA (miRNA) in 1993 (10), an increasing number of studies have revealed indispensable roles of miRNAs in tumorigenesis and/or cancer progression, as evidenced by the fact that approximately half of the miRNA genes are located in cancer-associated genomic regions or in fragile sites (11). miRNAs are short (18–25 nucleotide-long) and non-coding RNA molecules that regulate gene expression by suppressing mRNA translation and reducing mRNA stability, through imperfect complementary base pairing (12). There are two types of cancer-related miRNAs; oncogenic miRNAs (oncomiRs) and tumor-suppressive (TS) miRNAs. In general, oncomiRs are overexpressed in cancers while TS miRNAs are underexpressed. As the metabolism of cancer cells is highly regulated by signaling pathways related to oncogenes and tumor-suppressor genes (13), indispensable roles of miRNAs in the metabolic reprogramming of tumor cells have been studied extensively. For instance, glycolysis starts with the entry of glucose into cells via glucose transporters (GLUTs). Among those, GLUT1 has been the most widely studied and shown to be overexpressed in many cancers. A variety of miRNAs such as miR-132, miR-144, miR-148b, miR218, miR-340, and miR-451 have been shown to suppress GLUT1 expression, which leads to the inhibition of cell proliferation and/or enhancement of chemo-sensitivity (2,14-16). Hexokinase 2 is the first and rate-limiting enzyme in glucose metabolism that phosphorylates glucose to glucose-6-phosphate. miR-143 suppresses glucose consumption and lactate production, and miR-181b abrogates glycolysis by targeting hexokinase 2 (2). Pyruvate kinase M (PKM) is a rate-limiting glycolytic enzyme that regulates net ATP production, and PKM2 is highly expressed in proliferating cancer cells, promoting glycolysis. miR-122 and miR-124 were reported to inhibit PKM2 expression and accordingly suppress cancer metastasis and/or proliferation (2). Lactate dehydrogenase A is regulated by miRNAs such as miR-34a/c, miR-369-3p, miR-374a, and miR-4524a/b (2,17). miRNAs that regulate key enzymes associated with cancer cell metabolism are listed in Table 1.

Full table
In the recent article titled “miR-450a Acts as a Tumor Suppressor in Ovarian Cancer by Regulating Energy Metabolism”, Muys et al. focused on miR-450a as a tumor suppressor miRNA in ovarian cancer by regulating cancer energy metabolism (18). Ovarian cancer is the leading cause of death from gynecological malignancies among women. Global cancer statistics from 185 countries indicated 295,414 new cancer cases and 184,799 deaths due to this disease in 2018 (19). Considering its high degree of lethality, there is an urgent need to identify the underlying cause of disease and novel therapeutic options. In that sense, this is a pioneering study that has provided novel evidence of miRNA-targeting nuclear or mitochondrially encoded mitochondrial genes in ovarian cancer. The authors focused on miR-450a among more than 1,500 miRNAs reported, since several previous studies have shown that its expression was significantly downregulated in ovarian cancer tissues (20,21) and suggested its potential tumor suppressive role. First, they stated that overexpression of miR-450a in ovarian cancer cells reduced tumor growth in vitro and in vivo. RNA sequence analysis identified a panel of potential miR-450a targets (ACO2, TIMMDC1, MT-ND2, and ATP5B) which regulate energy metabolism. Using The Cancer Genome Atlas database (22), the expression levels of these genes were compared between ovarian tumors and normal tissue and found to be significantly reduced in tumors. They further revealed a significant negative correlation between ACO2 and ATP5B with miR-450a, suggesting that these are direct targets of miR-450a. Finally, they suggested a potential for miRNA replacement therapy with this miRNA. Transduction of miR-450a in ovarian cancer cell lines inhibited cancer invasion via dysregulation of epithelial-to-mesenchymal transition pathway genes such as vimentin, decreased the mitochondrial membrane potential and increased glucose uptake and viability after glutamine withdrawal, which are characteristics of less invasive ovarian cancer phenotype. Since miR-450a also inhibited ACO2, a key enzyme in the citric acid cycle, most glucose was probably metabolized directly into pyruvate. As the citric acid cycle is also important for glutamine metabolism, transduction of this miRNA decreased the glutaminolysis rate, which contributed to the decreased production of NADPH, lipids, amino acids, nucleic acids, and consequently signaling pathways associated with migration and invasion. Therefore, the authors proposed that replacement of miR-450a acts as a tumor suppressor by inhibiting key genes associated with cellular energy metabolism as well as epithelial-to-mesenchymal transition-related genes and could be a future candidate for ovarian cancer treatment.
Accumulating evidence suggests that the replacement of TS miRNAs or inhibition of oncomiRs might be used to modulate the activity of oncogenes that drive tumor progression. Currently, several preclinical trials investigating the role of miRNA therapy for cancer have been launched to establish novel therapeutic interventions. However, the outcomes of translational clinical trials for miRNA replacement therapy reported so far have been disappointing (23). Besides, its clinical application has been plagued by several unanswered questions, such as the selection of appropriate miRNAs for a particular tumor type, suitable delivery system, treatment velocity, and avoiding off-target effects (24). Since a single miRNA regulates hundreds of targets, a candidate miRNA that can control metabolism may affect other cellular processes, which could lead to unwanted side effects. Improving miRNA delivery systems by developing more appropriate carriers accompanied by the manipulation of epigenetic factors that enhance specific delivery to cancer tissues and stabilize miRNA function would be key clues for the future success of miRNA replacement therapy.
Acknowledgments
We thank Moe Matsui for providing secretarial assistance.
Funding: This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (No. 18K09227 to K Sawada).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Zois CE, Harris AL. Glycogen metabolism has a key role in the cancer microenvironment and provides new targets for cancer therapy. J Mol Med 2016;94:137-54. [Crossref] [PubMed]
- Subramaniam S, Jeet V, Clements JA, et al. Emergence of MicroRNAs as Key Players in Cancer Cell Metabolism. Clin Chem 2019;65:1090-101. [Crossref] [PubMed]
- Warburg O. On the origin of cancer cells. Science 1956;123:309-14. [Crossref] [PubMed]
- Marie SKN, Shinjo SMO. Metabolism and brain cancer. Clinics (Sao Paulo) 2011;66 Suppl 1:33-43. [Crossref] [PubMed]
- Wallace DC. Mitochondria and cancer Douglas. Nat Rev Cancer 2012;12:685-98. [Crossref] [PubMed]
- Vyas S, Zaganjor E, Haigis MC. Mitochondria and Cancer. Cell 2016;166:555-66. [Crossref] [PubMed]
- Pustylnikov S, Costabile F, Beghi S, et al. Targeting mitochondria in cancer: current concepts and immunotherapy approaches. Transl Res 2018;202:35-51. [Crossref] [PubMed]
- Nguyen TL, Durán RV. Glutamine metabolism in cancer therapy. Cancer Drug Resist 2018;1:126-38.
- Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci 2010;35:427-33. [Crossref] [PubMed]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [Crossref] [PubMed]
- Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene 2006;25:6220-7. [Crossref] [PubMed]
- Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res 2016;76:3666-70. [Crossref] [PubMed]
- Jang M, Kim SS, Lee J. Cancer cell metabolism: Implications for therapeutic targets. Exp Mol Med 2013;45:e45. [Crossref] [PubMed]
- Fan JY, Yang Y, Xie JY, et al. RETRACTED ARTICLE: MicroRNA-144 mediates metabolic shift in ovarian cancer cells by directly targeting Glut1. Tumour Biol 2016;37:6855-60. [Crossref] [PubMed]
- Ding X, Liu J, Liu T, et al. miR-148b inhibits glycolysis in gastric cancer through targeting SLC2A1. Cancer Med 2017;6:1301-10. [Crossref] [PubMed]
- Li P, Yang X, Cheng Y, et al. MicroRNA-218 Increases the Sensitivity of Bladder Cancer to Cisplatin by Targeting Glut1. Cell Physiol Biochem 2017;41:921-32. [Crossref] [PubMed]
- Wang J, Wang H, Liu A, et al. Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget 2015;6:19456-68. [PubMed]
- Muys BR, Sousa JF, Placa JR, et al. MiR-450a acts as a tumor suppressor in ovarian cancer by regulating energy metabolism. Cancer Res 2019;79:3294-305. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Zhang L, Volinia S, Bonome T, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A 2008;105:7004-9. [Crossref] [PubMed]
- Li Y, Yao L, Liu F, et al. Characterization of microRNA expression in serous ovarian carcinoma. Int J Mol Med 2014;34:491-8. [Crossref] [PubMed]
- National Cancer Institute. The Cancer Genome Atlas Program [cited 2019 Oct 22]. Available online: http://cancergenome.nih.gov/
- Hosseinahli N, Aghapour M, Duijf PHG, et al. Treating cancer with microRNA replacement therapy: A literature review. J Cell Physiol 2018;233:5574-88. [Crossref] [PubMed]
- Subramaniam S, Jeet V, Clements JA, et al. Emergence of MicroRNAs as Key Players in Cancer Cell Metabolism. Clin Chem 2019;65:1090-101. [Crossref] [PubMed]


