Matrix-assisted laser desorption ionization time-of-flight mass spectrometry combined with UF-5000i urine flow cytometry to directly identify pathogens in clinical urine specimens within 1 hour
Introduction
Urinary tract infections (UTIs) caused by abnormal growth and reproduction of various pathogenic microorganisms in the urinary tract is one of the most common infectious diseases in outpatients and inpatients (1). It is reported that about 150 million people suffer from UTI every year in the world, and the cost of treatment for UTI exceeds 6 billion US dollars (2). The diagnosis of UTI includes clinical manifestations and laboratory tests, and the definite diagnosis was based on urine culture (3). However, the method requires 24 to 48 hours, and approximately 80% of urine cultures are negative (4), which can’t provide pathogen evidence for diagnosis in time. matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a new tool for microbial detection. It has the advantages of fast, accurate, low cost and simple operation, and achieved considerable results in the identification of bacteria (5). UF-5000i urine flow cytometry, as a new technology for clinical UTIs screening in recent years, has play a important role in the early screening of UTIs. In this study, to explore the application of MALDI-TOF MS and UF-5000i in the etiological diagnosis of UTIs, we introduced a new method to directly identify pathogens in urine specimens by MALDI-TOF MS and UF-5000i to save the pathogen diagnosis time of UTIs, meanwhile, conventional urine culture and bacterial identification by MALDI-TOF MS were used as the gold standard for comparison.
Methods
Ethical approval
The collection of all samples was approved by the Affiliated Hospital of Xuzhou Medical University and was obtained after the patient agreed to the research purpose. The consent procedure was also approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University. IRB approval number was XYFY2019-KL110-01.
Urine sample collection and proposed protocol design
We collected 1,503 urine samples submitted to the Clinical Microbiology Laboratory, Department of Clinical Laboratory, Affiliated Hospital of Xuzhou Medical University (Xuzhou, Jiangsu, China) during August 2018 to January 2019 from inpatients and outpatients with symptoms of UTIs. Each volume of urine sample was not less than 20 mL, which was divided into three parts using sterile centrifuge tubes. The first 5 mL liquid was used for conventional cleaning mid-stream urine culture, the second 5 mL liquid for UF-5000i (Sysmex, Kobe, Japan) analysis to screen out the bacterial counts, which were more than 1×105 bacteria/mL. The last 10 mL liquid was subsequently processed to prepare bacterial pellets and was directly identified by the MALDI-TOF MS (Bruker Daltonik, GmbH, Bremen, Germany).
Quality control strains
Escherichia coli was ATCC25922, Klebsiella pneumoniae was ATCC13883, Pseudomonas aeruginosa was ATCC27853, Staphylococcus aureus was ATCC25923, Enterococcus faecalis was ATCC29212 and Proteus mirabilis was ATCC3563.
Routine protocol processing
The routine protocol included conventional cleaning mid-stream urine culture and bacterial identification by MALDI-TOF MS: all the prepared urine samples were performed by a fully automated plating and streaking equipment (DIASE, GmbH, Wuhan, China). In detail, the first portion with 5 mL of urine was processed by DIASE using 10 microlitre loops on blood agar and MacConkey agar plates for culture. The plates were incubated for 24 to 48 h at 35 °C and 5% CO2 concentrations.
One or two significant UTI pathogens grow on the culture, and ≥104 CFU/mL were considered to be positive urine specimens. Meanwhile, three or more microorganisms were considered contaminated. At last, MALDI-TOF MS was used for pathogens identification.
UF-5000i urine flow cytometry screening
The second portion with 5 mL urine was used for UF-5000i (Sysmex, Kobe, Japan) automated urine flow cytometry to screen out the bacterial counts. And samples with a cut-off of 1×105 bacteria/mL were further selected and processed by MALDI-TOF MS for pathogens detection.
Urine sample pretreatment and bacterial purification
The third portion with 10 mL urine sample was placed in 15 mL sterile centrifuge tube, centrifuged at 1,000 ×g for 1 min to remove cells and other impurities; the supernatant was then collected and centrifuged at 15,500 ×g for 5 min to collect bacteria pellet. After that, the supernatant was discarded. The pellet was resuspended in 1 mL of 0.9% saline solution and transferred into a 2 mL micro-centrifuge tube. Next, the pellet was centrifuged at 18,500 ×g for 5 min (6). Finally, the bacterial purification had been accomplished.
Direct identification of bacteria by MALDI-TOF MS
The purified bacterial pellet of the third part was agitated by oscillator (Kanghe, Shanghai, China) for 30 s, then, 2 µL of the supernatant from the third portion directly spotted onto a polished steel target plate. Dried spots were g overlaid with 2 µL 70% formic acid. After air-dried at room temperature, 2 µL of α-cyano-4-hydroxycinnamic acid (HCCA) matrix solution (Bruker Daltonik, GmbH, Bremen, Germany) was added. After drying of the plate, MALDI-TOF MS measurements were performed with a Microflex LT benchtop mass spectrometer (Bruker Daltonik, GmbH, Bremen, Germany) equipped with a 60 Hz nitrogen laser. Parameter settings have been optimized for the appropriate (MBT_FC) mass range (ion source IS1, 20 kV; IS2, 18.0 kV; lens, 6 kV; detector gain, 2,689 V; gating, none). Spectra were recorded in the positive linear mode in the mass range of 2,000 Da to 20,000 Da with the maximum laser frequency. Analysis of the spectra was applied by MALDI Biotyper software (Bruker Daltonics) and Flex Analysis. According to the pattern recognition algorithm of peak position, peak intensity distribution and peak frequency, the measured spectra were compared with the reference biotyper library. Refer to the manufacturer, a score of ≥2.0 suggests credible species-level identification, a score between 1.7 and 1.99 suggests credible genus-level identification, and a score of <1.7 indicates that the identification result is not credible. According to these criteria, the results of direct identification by MALDI-TOF MS were compared with those of routine protocol for evaluation of proposed protocol accuracy.
Results
A total of 1,503 cases of clean mid-stream urine samples were screened by UF-5000i, Then conventional bacterial culture identification and MALDI-TOF MS direct identification were performed separately. The specific results are as follows.
The cut-off value of UF-5000i
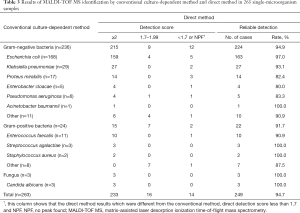
In order to identify the cut-off value of UF-5000i, we performed a pilot study to determine the minimum amount of bacteria required for MALDI-TOF MS identification of the major pathogens in UTIs. The results are shown in Table 1, It is suggested that: Reliable direct identification (genus-level) by MALDI-TOF MS revealed that the lowest bacterial count of Escherichia coli was 6×104/mL, the lowest bacterial count of Pseudomonas aeruginosa was 1×105/mL, the lowest bacterial count of Staphylococcus aureus was 1×105/mL, and the lowest bacterial count of Enterococcus faecalis was 3×105/mL.

Full table
To get reliable results, in the direct identification of urine specimens by MALDI-TOF MS, Gram-positive cocci need more bacteria than gram-negative bacilli. In other words, in UTIs, the direct identification of gram-positive cocci is more difficult than that of gram-negative bacilli. Therefore, for the identification of gram-positive cocci, we can collect the bacterial by increasing the centrifugal urine volume (7). On the other hand, diagnostic criteria for UTIs according to the American Association of Infectious Diseases, in patients with symptomatic lower UTI, the number of bacterial culture colonies in fresh clean midstream urine is ≥103 CFU/mL, upper UTI (pyelonephritis), quantitative bacterial culture colonies number ≥104 CFU/mL. And Hooton et al. (8) proposed the determination of asymptomatic bacteriuria: the same bacteria in the midstream urine culture for 2 consecutive and the number of colonies ≥105 CFU/mL. Finally, through preliminary experiments, we determine the cut-off value was 1×105 bacterial/mL.
UF-5000i urine flow cytometry detection
During the test period, 1,503 urine specimens submitted to our clinical humoral laboratory for pilot study by UF-5000i. Based on the formerly established bacterial pellets counts cut-off value of ≥105 bacteria/mL, 296 out of 1,503 specimens were be regarded as positive for further analysis (Figure 1).
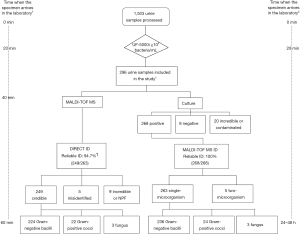
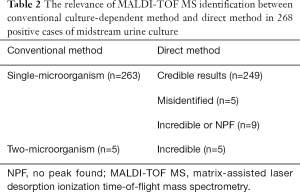
Conventional urine culture and bacterial identification
Among 296 selected urine samples, which were screened by UF-5000i; 263 had significant growth of single-microorganism by urine culture, 5 had significant growth of two-microorganism, 20 were unreliable or contaminated and 8 were negative (Table 2, Figure 1). Among 263 single-microorganism, Gram-negative bacteria were 236, included 168 Escherichia coli, 29 Klebsiella pneumoniae, 17 Proteus mirabilis, 5 Enterobacter cloacae, 6 Pseudomonas aeruginosa and 12 other species. In the midst of 24 Gram-positive bacteria, including 11 Enterococcus faecalis, 3 Streptococcus agalactiae, 2 Staphylococcus aureus and 8 other species. Then, among 3 fungus, both of them were Candida albicans (Table 3).
Full table
Full table
MALDI-TOF MS direct identification
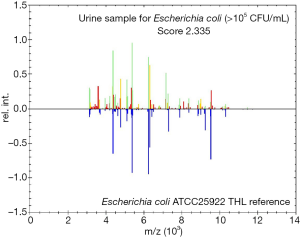
In this experiment, 296 of 1,503 bacterial pellets from urine specimen were directly identified by the MALDI-TOF MS. Compared with the conventional culture-dependent method, 249 (249/263, 94.7%) results in keeping with it, and they were both single-microorganism, 5 samples were misidentified (Table 2) and 42 results were unreliable (data not shown). Among 249 credible results, species-level identification (score ≥2.0) was contained 233 (233/249, 93.6%) cases, 16 (16/249, 6.4%) samples scored between 1.7 and 1.99. Meanwhile, Gram-negative bacteria results were 224, included 163 Escherichia coli, 27 Klebsiella pneumoniae, 14 Proteus mirabilis, 4 Enterobacter cloacae, 5 Pseudomonas aeruginosa and 11 other species. In the 22 Gram-positive bacteria, including 10 Enterococcus faecalis, 3 Streptococcus agalactiae, 2 Staphylococcus aureus and 7 other species. The rest of 3 fungus were both Candida albicans (Table 3). MALDI-TOF MS directly identified Escherichia coli (UF-5000i bacterial count >105/mL) in the mid-stream urine specimens and compared it with the best matching spectrum in the Biotyper database, Green indicated good matching peaks, yellow indicated a moderate match, and red indicated no match (Figure 2).
Discussion
Recently, In the clinical microbiology laboratory, MALDI-TOF MS had made considerable achievements in microbial identification, especially in the field of bacteria and fungi (9,10). And for common clinical bacteria, MALDI-TOF MS can accurately identify 84.1–93.6% of the bacteria to the species level (11). Moreover, MALDI-TOF MS has a high degree of stability and accuracy in the identification of bacterial based on traditional culture, which greatly shortens the time of conventional bacterial species identification (12,13), and its cost is lower than routine methods, such as BD Phoenix systems, Vitek2-Compact bacterial identification system, etc (14,15).
UF-1000i automatic urine flow cytometry is an advanced urine tangible component analysis system. Since its launch in 2005, it has been popular with clinical workers for its convenient, fast and efficiency (16). UF-1000i has a specialized bacterial detection channel that can accurately provide the bacterial count from the urine sample (17). UF-5000i is a new urine flow cytometry developed by Sysmex Corporation in recent years, which is generally the same as UF-1000i in terms of bacterial detection. UF-5000i can’t only count the amount of bacteria in urine samples, and it has an advantage in distinguishing Gram-positive bacteria, Gram-negative bacteria and mixed bacteria. The information on bacterial Gram staining is inferred from the scatter plot. Our experiment also did some research on the urine scatter plot. It can be proved that this technology is credible for the initial identification of Gram-negative bacilli and Gram-positive cocci. But the results were not shown. Therefore, UF-5000i plays a major role in the initial screening of UTIs.
In this study, the combination of MALDI-TOF MS and UF-5000i was used to directly identify the pathogenic bacteria in urine specimens in a rapid and accurate method, and the preliminary test results could be provided within 1 hour, while the identification time required for conventional urine culture was 18–24 h (4). This method reduces the identification time of pathogenic bacteria and is of great significance in the improvement of methodology in clinical microbiological laboratory. Its speed and relatively low cost are likely to replace traditional biochemical identification and automated bacterial identification instruments, providing an effective basis for the diagnosis of UTIs. The method can even reduce unnecessary empirical antibiotic treatment, so as to target drugs based on early detection of pathogens, thereby decreasing bacterial resistance and side effects (18).
The results of this study showed that in 268 specimens with positive urine culture, MALDI-TOF MS directly identified 249 (249/263, 94.7%) cases (above genus level) consistent with the results of traditional methods. The coincidence rate is slightly higher than that of Wang et al. (19). Maybe it’s because, In the pretreatment of urine specimens, the centrifugal speed has been improved, and the operation process has been streamlined. These 263 reliable identifications are both single colonies. Based on these data, we can infer that the main pathogens of UTIs are still Gram-negative bacilli, and the dominant one is Escherichia coli, which is consistent with reports in the literature (20). Of the 236 Gram-negative bacilli, 9 (9/236, 3.8%) were identified as genus level and 225 (225/236, 95.3%) were identified as species level; Of the 24 Gram-positive cocci, 7 (7/24, 29.2%) were identified as genus level and 15 (15/24, 62.5%) were identified as species level. It can be inferred that the identification effect of Gram-negative bacilli is better than that of Gram-positive cocci (21). By UF-5000i urine flow cytometry screening, Gram-negative bacilli counts significantly higher than Gram-positive cocci, which may be due to the suitable growth environment of Gram-negative bacilli in urine, resulting in rapid reproduction (20).
Regarding the detection of fungi, Íñigo et al. (6) believed that identification of yeasts from direct urine samples was still unresolved. However, in our experiments, 3 fungi were reliably identified, and the coincidence rate of MALDI-TOF MS direct method identification was 100.0%. Refer to a study recently by Galán et al., In urine, vaginal discharge, sputum and blood culture, 94.8% of the yeasts isolated were correctly identified (22), and in the study by Wattal et al., the recognition rate of yeast by MALDI-TOF MS was 92.8%, and that of filamentous fungi was 80% (23). The reason for the high coincidence rate may be that the MALDI-TOF MS uses a formic acid extraction method to identify fungi (24), and formic acid can lyse the cytoderm of the fungus to fully expose the protein component for detection purposes.
In 5 urine samples from which two microorganisms were cultured, the MALDI-TOF MS direct identification method was unreliable. It can be seen that MALDI-TOF MS does not provide reliable identification results for identifying mixed infections of two species of bacteria. It has been reported in the literature that the identification of MALDI-TOF MS in mixed-growth urine culture may depend on the ratio between the two bacteria; and when the two bacteria are mixed, MALDI-TOF MS can only identify one of the dominant bacteria. When the ratio of these two kinds of bacteria is similar, the identification rate will be lower or even failure (21). In addition to the mixed infection of the two kinds of bacteria will interfere with the identification of urinary pathogens, there are still some other influencing factors, such as the number of bacteria in the urine, cellular components, protein, urobilinogen and other impurities. For pre-treatment of urine, Veron et al. compared the performance of differential centrifugation, urine filtration and short culture in the identification of uropathogens by MALDI-TOF MS. The correct recognition rate of MALDI-TOF MS is higher by the urine filtration and the short culture method (25). Therefore, the pre-processing of specimens is particularly important. Besides, we can also determine whether there is a mixed bacterium in the urine specimen through the bacterial channel of UF-5000i, so that we can know whether the sample can be directly identified by MALDI-TOF MS (17).
Gold can’t be pure and man can’t be perfect. Our experimental methods still have some limitations. For example, this method requires at least 20 mL of the original urine specimen, the amount of bacteria in the urine can’t be lower than 1×105/mL. Moreover, it is unable to provide reliable identification of UTIs caused by multiple pathogenic microorganisms. Fortunately, UTIs caused by a single microorganism still account for the vast majority, and a sufficient volume of original urine specimens are more easily obtained in clinical practice. However, one of the most obvious drawbacks is that the method does not provide results from antimicrobial susceptibility testing. Thus, traditional methods based on culture are necessary. Oviaño et al. used imipenem as an antibiotic marker to directly detect carbapenem resistance in the original urine specimen and the results were automatically analyzed using the STARBL module of MALDI-TOF Biotyper Compass software (Bruker Daltonik, Germany) (26). Angaali Neelima et al. used VITEK 2 COMPACT automatic microbial identification system for direct identification and sensitivity test of Gram-negative bacilli in urine samples (27). Mirande et al. used faropenem and ertapenem hydrolysis as a model to compare the benefits and weaknesses of rapid detection of carbapenemase activity by MALDI-TOF MS (28). Therefore, the susceptibility test of urine specimens without relying on culture will be our next research direction.
Conclusions
MALDI-TOF MS combined with UF-5000i Urine Flow Cytometry to identify the pathogenic bacteria in urine directly, as a fast and innovative method, saves at least 23 hours, provides a reliable and efficient etiology basis for the diagnosis of UTIs timely.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81871734); Jiangsu Provincial Medical Talent (ZDRCA2016053); Six Talent Peaks Project of Jiangsu Province (WSN-135) and Advanced Health Talent of Six-One Project of Jiangsu Province (LGY2016042).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Zhi-De Hu, Bing Gu) for the series “Advances in laboratory tests for infectious diseases” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2019.10.73). The series “Advances in Laboratory Tests for Infectious Diseases” was commissioned by the editorial office without any funding or sponsorship. BG served as the unpaid Guest Editor of the series and serves as an unpaid Managing Editor of Annals of Translational Medicine. The other authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University (IRB: XYFY2019-KL110-01) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Stamm WE, Norrby SR. Urinary Tract Infections: Disease Panorama and Challenges. J Infect Dis 2001;183 Suppl 1:S1-4. [Crossref] [PubMed]
- Akoachere JF, Yvonne S, Akum NH, et al. Etiologic profile and antimicrobial susceptibility of community-acquired urinary tract infection in two Cameroonian towns. BMC Res Notes 2012;5:219. [Crossref] [PubMed]
- De Cueto M, Aliaga L, Alós JI, et al. Executive summary of the diagnosis and treatment of urinary tract infection: Guidelines of the Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC). Enferm Infecc Microbiol Clin 2017;35:314-20. [Crossref] [PubMed]
- Patel HD, Livsey SA, Swann RA, et al. Can urine dipstick testing for urinary tract infection at point of care reduce laboratory workload? J Clin Pathol 2005;58:951-4. [Crossref] [PubMed]
- Seng P, Drancourt M, Gouriet F, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 2009;49:543-51. [Crossref] [PubMed]
- Íñigo M, Coello A, Fernández-Rivas G, et al. Direct Identification of Urinary Tract Pathogens from Urine Samples, Combining Urine Screening Methods and Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J Clin Microbiol 2016;54:988-993. [Crossref] [PubMed]
- Demarco ML, Burnham CA. Diafiltration maldi-tof mass spectrometry method for culture-independent detection and identification of pathogens directly from urine specimens. Am J Clin Pathol 2014;141:204-12. [Crossref] [PubMed]
- Hooton TM, Scholes D, Stapleton AE, et al. A prospective study of asymptomatic bacteriuria in sexually active young women. N Engl J Med 2000;343:992-7. [Crossref] [PubMed]
- Grosse-Herrenthey A, Maier T, Gessler F, et al. Challenging the problem of clostridial identification with matrix-assisted laser desorption and ionization–time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe 2008;14:242-9. [Crossref] [PubMed]
- Marklein G, Josten M, Klanke U, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J Clin Microbiol 2009;47:2912-7. [Crossref] [PubMed]
- Bizzini A, Greub G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect 2010;16:1614-9. [Crossref] [PubMed]
- Carbonnelle E, Mesquita C, Bille E, et al. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin Biochem 2011;44:104-9. [Crossref] [PubMed]
- Neville SA, Lecordier A, Ziochos H, et al. Utility of matrix-assisted laser desorption ionization-time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J Clin Microbiol 2011;49:2980-4. [Crossref] [PubMed]
- Yan Y, Meng S, Bian D, et al. Comparative evaluation of Bruker Biotyper and BD Phoenix systems for identification of bacterial pathogens associated with urinary tract infections. J Clin Microbiol 2011;49:3936-9. [Crossref] [PubMed]
- Tan KE, Ellis BC, Lee R, et al. Prospective evaluation of a matrix-assisted laser desorption ionization-time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol 2012;50:3301-8. [Crossref] [PubMed]
- Jolkkonen S, Paattiniemi EP, Sarkkinen H. Screening of urine samples by flow cytometry reduces the need for culture. J Clin Microbiol 2010;48:3117. [Crossref] [PubMed]
- Jiang T, Chen P, Ouyang J, et al. Urine particles analysis: performance evaluation of Sysmex UF-1000i and comparison among urine flow cytometry, dipstick, and visual microscopic examination. Scand J Clin Lab Invest 2011;71:30-37. [Crossref] [PubMed]
- Harbarth S, Garbino J, Pugin J, et al. Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immunomodulating therapy for severe sepsis. Am J Med 2003;115:529-35. [Crossref] [PubMed]
- Wang XH, Zhang G, Fan YY, et al. Direct identification of bacteria causing urinary tract infections by combining matrix-assisted laser desorption ionization-time of flight mass spectrometry with UF-1000i urine flow cytometry. J Microbiol Methods 2013;92:231-5. [Crossref] [PubMed]
- Al-Naqshbandi AA, Chawsheen MA, Abdulqader HH. Prevalence and antimicrobial susceptibility of bacterial pathogens isolated from urine specimens received in rizgary hospital-Erbil. J Infect Public Health 2019;12:330-6. [Crossref] [PubMed]
- Ferreira L, Sanchez-Juanes F, Gonzalez-Avila M, et al. Direct Identification of Urinary Tract Pathogens from Urine Samples by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J Clin Microbiol 2010;48:2110-5. [Crossref] [PubMed]
- Galán F, García-Agudo L, Guerrero I, et al. Evaluation of mass spectrometry for the identification of clinically interesting yeast. Enferm Infecc Microbiol Clin 2015;33:372-8. [PubMed]
- Wattal C, Oberoi JK, Goel N, et al. Matrix-assisted laser desorption ionization time of flight mass spectrometry (maldi-tof ms) for rapid identification of micro-organisms in the routine clinical microbiology laboratory. Eur J Clin Microbiol Infect Dis 2017;36:807-12. [Crossref] [PubMed]
- Lee HS, Shin JH, Choi MJ, et al. Comparison of the Bruker Biotyper and VITEK MS Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry Systems Using a Formic Acid Extraction Method to Identify Common and Uncommon Yeast Isolates. Ann Lab Med 2017;37:223-30. [Crossref] [PubMed]
- Veron L, Mailler S, Girard V, et al. Rapid urine preparation prior to identification of uropathogens by MALDI-TOF MS. Eur J Clin Microbiol Infect Dis 2015;34:1787-95. [Crossref] [PubMed]
- Oviaño M, Ramírez CL, Barbeyto LP, et al. Rapid direct detection of carbapenemase-producing Enterobacteriaceae in clinical urine samples by MALDI-TOF MS analysis. J Antimicrob Chemother 2017;72:1350-4. [Crossref] [PubMed]
- Angaali N, Vemu L, Padmasri C, et al. Direct identification and susceptibility testing of Gram-negative bacilli from turbid urine samples using VITEK2. J Lab Physicians 2018;10:299-303. [Crossref] [PubMed]
- Mirande C, Canard I, Buffet Croix Blanche S, et al. Rapid detection of carbapenemase activity: benefits and weaknesses of MALDI-TOF MS. Eur J Clin Microbiol Infect Dis 2015;34:2225-34. [Crossref] [PubMed]