
Do lncRNAs and circRNAs expression profiles influence discoid lupus erythematosus progression?—a comprehensive analysis
Introduction
As a chronic inflammatory cutaneous disease, discoid lupus erythematosus (DLE), unless diagnosed and treated in a timely fashion, can lead to disfiguring scarring and skin atrophy (1,2). DLE lesions occur in approximately 6–10% of patients with systemic lupus erythematosus (SLE) (3,4). Although there are similarities in histology, their clinical course and prognosis are different, suggesting different pathogeneses (5). DLE’s pathophysiology has not been extensively investigated and is likely related to a complex of environmental, genetic, and immune cell interactions. Recent studies have found that dendritic cells, natural killer cells, and toll-like receptors play a dominant role in this process. Other evidence has also shown that, compared with healthy and involved DLE skin, there are several signaling pathways related to T helper (Th) type 1 in lesional DLE (4). However, the understanding of key pathogenic pathways is far from clear.
An increasing number of high-throughput deep sequencing results have found that only a small proportion of the human genome is transcribed into protein-coding mRNAs, while most genome is transcribed into noncoding RNAs (ncRNAs) (6-8), which are considered a new type of long noncoding RNA (lncRNA) with a length of more than 200 nucleotides (9-11). Recent evidences suggest that lncRNAs are involved many pathophysiological mechanisms, and are frequently deregulated (12-14) in autoimmune thyroid disease and rheumatoid arthritis (RA) (9). However, the functional and genomic profile of lncRNAs in DLE remains unclear. As a new topic in scientific literature, noncoding and circular RNAs (circRNAs) with a covalently closed loop structure (15,16) have been already mostly described in eukaryotic cells (17). They can occur in any part of genome and regulate gene expression (17,18). In recent years, circRNAs have been found to have numerous functional microRNA binding sites and occupy the role of miRNA regulators of gene expression. Accumulating evidence show that circRNAs are part of various physiological and pathological conditions, including cancer, prion infection, neurological disorders, and atherosclerotic vascular diseases (17-21). Despite this knowledge, little is known about the functional roles of circRNA in DLE, and the expression profiles of the noncoding RNAs role in the progression of DLE remain elusive.
We therefore evaluated the differential pattern expression of lncRNA/mRNA/circRNA in order to explore the molecular profiles of non-coding RNAs in DLE by high-throughput RNA-sequencing (RNA-seq). We initially analyzed their pathways, Gene Ontology (GO) items, functional coding-noncoding co-expression networks, and TF-lncRNA-enriched mRNA networks. Our results from this analysis demonstrate that numerous lncRNAs and circRNAs have potential to be considered diagnostic and therapeutic markers for DLE. Finally, the functions of the lncRNAs and circRNAs were predicted by evaluating the differences of expression in lncRNA/circRNA/mRNA co-expression networks. These results provide further insight into the underlying pathogenesis of DLE and offer targets for DLE therapy.
Methods
Patients and lower lip samples
After written consent was given, a punch biopsy of 6 mm was performed in each of the 3 patients affected by DLE from the lesional and non-lesional lower lips. All samples were evaluated by histology. The local ethics committee of Shanghai Ninth People’s Hospital approved this study.
RNA collection, library preparation, and sequencing
Total RNAs of DLE and normal control samples were extracted using the TriZol Isolation Kit (Ambion). The libraries were constructed and sequenced on the Illumina sequencing platform (HiSeqTM 2500 or other platforms) according to the manufacturer’s instructions.
Differential expression
The statistical significance of differentially expressed lncRNAs/circRNAs/mRNAs between the DLE group and normal control group was identified by P value and FDR filtering (fold change ≥2.0, P<0.05, and FDR <0.05). The overview of the characteristics of expression profiles was shown by hierarchical clustering.
Correlation analysis
The Pearson correlation coefficient (PCC ≥0.90, P value <0.01, and FDR <0.01) between genes was calculated to perform the co-expression analysis according to their expression levels.
GO and pathway analysis
GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed as previously described.
Cis and trans regulation prediction
As previously described (22), mRNAs co-expressed with lncRNAs which significantly overlapped with the target genes of a given transcription factor were enriched, and the lncRNAs-TF mRNA network was established.
Results
The expression of mRNA, lncRNA, and circRNA profiles
Thousands of transcripts were detected by the RNA-seq in DLE and adjacent normal tissues (Figure 1). A total of 555 mRNAs (Figure 1A) and 507 lncRNAs (Figure 1B) were found to be differentially expressed with fold change ≥2.0, P<0.05, and FDR <0.05. Among them, 204 and 351 mRNAs were upregulated and downregulated, respectively. A total of 198 and 309 lncRNAs were upregulated and downregulated in 3 DLE tissues compared with adjacent normal controls, respectively. Moreover, filtering analysis (fold change ≥2.0, P<0.05. and FDR <0.05) identified a total of 62 (30 up-regulated and 32 down-regulated) differentially expressed circRNAs (Figure 1C) in DLE compared with normal tissues. Furthermore, a total of 161 lncRNAs showed fold change ≥10 (up: 57, down: 104). HCP5 was the most upregulated lncRNA in DLE with a fold change of almost 206, compared with the adjacent normal tissue. Hierarchical clustering demonstrated that mRNA, lncRNA, and circRNA expression patterns among DLE tissues were distinguishable and different from those in the matched normal tissues.
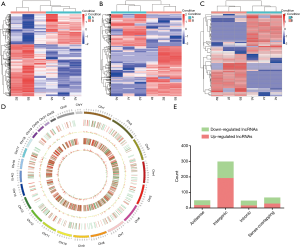
Accordingly, the detected lncRNAs were widely distributed in all chromosomes including the sex chromosomes X and Y (Figure 1D). According to their relation with protein-coding genes, the lncRNAs were divided into four categories (Figure 1E): antisense, sense overlapping, intergenic, intronic notably. Intergenic lncRNAs were the largest category (65.0%). From further analysis, the percentages of intergenic lncRNAs in up-regulated and down-regulated lncRNAs in DLE compared with controls was 76.4% and 51.2%, respectively.
GO and KEGG pathway analysis
GO enrichment analysis of differentially expressed mRNAs indicated their role in dramatically regulating lncRNAs. Our data showed that the up-regulated mRNAs associated with biological processes, cellular components and molecular function, were immune response, integral components of plasma membrane, and chemokine activity, respectively (Figure 2A). Meanwhile, the down-regulated transcripts were most related to retina homeostasis, apical plasma membrane, and ligand-gated sodium channel activity (Figure 2B). Notably, based on GO terms, the immune and inflammatory response is

KEGG pathway enrichment analysis of differentially expressed mRNAs was performed to reveal the pathways and molecular interactions related to genes. We found 20 pathways associated with upregulated mRNAs and 20 related to downregulated mRNAs in the KEGG pathway enrichment analysis. As shown, the staphylococcus aureus infection and viral myocarditis signaling pathway were the top pathways in upregulated protein-coding genes (Figure 2C), whereas the top enriched KEGG pathway was aldosterone-regulated sodium reabsorption and salivary secretion for downregulated transcripts (Figure 2D). The result suggests that these pathways might be critical to the progression of DLE.
To determine whether circRNAs regulate the transcription of parental genes, the genes producing differently expressed circRNAs were divided by GO analysis. Compared to adjacent normal tissues, the gene expression profile of linear counterparts of differentially up-regulated circRNAs in the DLE group favored protein ubiquitination (biological progress), golgi membrane (cellular component) and transcription factor activity, and sequence-specific DNA binding (molecular function) (Figure 2E). Meanwhile, GO enrichment analysis of downregulated transcripts showed that the closely related GO terms were intracellular signal transduction (biological progress), apical plasma membrane (cellular component), and RNA polymerase II regulatory region sequence-specific DNA binding (molecular function) (Figure 2F). These results suggest that these molecular functions and biological process could be involved in DLE progression.
lncRNA/mRNA expression and function
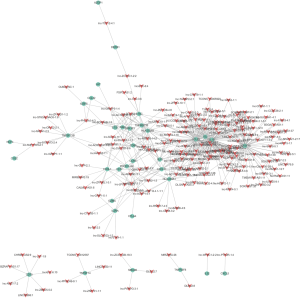
As the functionality of most lncRNAs has thus far not been annotated, the functional prediction of lncRNAs was based on the annotation of the co-expressed mRNA function. To build a CNC network, we chose 37 significantly expressed coding genes in the DLE group according to the degree of correlation (Figure 3). The mRNAs were involved in several biological processes, including immune response, immunological synapse, and T cell activation. The network indicated that up-regulated lncRNA lnc-MIPOL1-6 was positively correlated, and down-regulated lncRNA lnc-DDX47-3 was negatively correlated with IL19, CXCL1, CXCL11, and TNFSF15, which are involved in immune response. The co-expression network suggested that mRNA or lncRNA might be associated with one to dozens of lncRNAs, and the regulation between lncRNAs and mRNAs is implicated in DLE.
Cis and the trans-regulating function prediction of lncRNAs
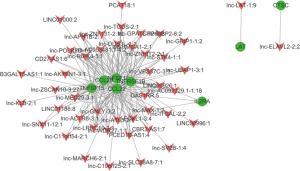
We further analyzed how the dysregulated mRNAs might play a cis or trans-regulatory role in lncRNA genes according to co-expression. We then built a correlated expression network to identify the relation between the 8 mRNAs and their adjacent coding gene. The 8 differentially expressed mRNAs were chosen to hunt their nearby coding genes. The co-expressed lncRNAs genes were defend as co-regulated genes with one differentially expressed mRNA on the same chromosome within 300 kb (Figure 4). This network could provide valuable clues about the mRNAs adjacent to the lncRNAs genes.
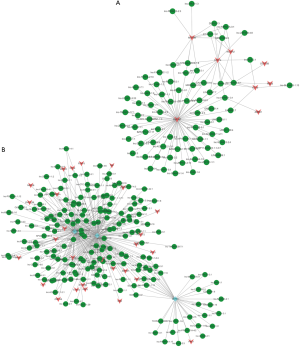
To further evaluate the role of lncRNAs in DLE, according to the enrichment with cumulative hypergeometric test, we searched the TFs correlated with lncRNAs and constructed a co-expression network by combining differentially expressed lncRNAs with TFs. We then predicted the trans-regulatory functions of lncRNAs by the TFs that could regulate their expression. Some lncRNAs might be involved in particular pathways regulated by TFs, supposing lncRNAs could have trans-regulatory functions. Therefore, we analyzed the co-expressed mRNAs with these TF-regulated mRNAs and lncRNAs. With the threshold of FDR <0.01 and P<0.01, each lncRNA could connect with one to a dozen TFs and each pair of lncRNA-TF resulted in the enrichment of several genes (Figure 5A)—a discovery which may provide critical data for subsequent research. As shown in Figure 4A, dysregulated lncRNAs were found to correspond to 8 TFs. Next, we further introduced mRNAs to build the ternary network of TF-lncRNA-mRNA base on TF-lncRNA binary analyses (Figure 5B). The results demonstrate that most of the lncRNAs participated in pathways regulated by TFs (STAT4, ETV6, and ZNF597), suggesting that these TFs could be correlated with the pathogenesis and development of DLE.
Discussion
DLE is the most common form of cutaneous lupus erythematosus, accounting for approximately 80% of cases (1,4). Traditional research in gene regulation has focused on protein-coding genes, and a large number of non-coding RNAs including lncRNAs and circRNAs have been described, (9,19,22). In particular, emerging evidence of deregulated lncRNA expression covering SLE (23-26) has implied that abnormal lncRNA expression may be involved in the progression of DLE. Recently, circRNAs were also reported to have a potential role in SLE (27-30). Thus far, however, a systematic analysis of noncoding RNA (lncRNAs and circRNAs) differential expression profiles in DLE has not been conducted. To further identify the functions in DLE, we evaluated lncRNA and circRNA expression profiles in the genome of 3 patients affected by DLE and matched adjacent tissues using high-throughout sequencing.
We characterized lncRNA and circRNA expression using RNA-seq. A total of 507 (up: 198, down: 309) lncRNAs and 161 (up: 57, down: 104) circRNAs showed significant differential expression in DLE. Most of the deregulated lncRNAs (65%) belonged to the intergenic category. We further found that the differential expression of lncRNAs was highlighted on each chromosome, suggesting that each chromosome was associated with different degrees of abnormality in DLE progression. As reported previously, our data also revealed a 13-fold up-regulation of macrophage migration inhibitory factor (MIF) in DLE. It was been shown that MIF has a crucial role as a regulator in acute and chronic immuno-inflammatory conditions like atherosclerosis, RA, and more recently, SLE (31,32). Of note, our data also showed that a 205-fold increase of lncRNA HCP5 in DLE, which was also reported to be associated with disease development in SLE (33). Other dysregulated lncRNAs of the present study were also found and verified in other studies. However, most of the differentially expressed lncRNAs are reported here for the first time.
Compared with the adjacent normal controls, the most significant annotation results which were derived from GO items were immune response, inflammatory response, and T cell co-stimulation in the DLE group, suggesting that deregulated mRNAs may play a crucial role in the pathways of regulation for DLE. KEGG pathway analysis revealed that 20 pathways could contribute to the pathogenesis of DLE including staphylococcus aureus infection and viral myocarditis signaling pathway. This further suggests that the dysregulated activation of the immune system function is strongly correlative with DLE.
With the construction of the CNC co-expression network, many lncRNA expression levels were found to be associated with the expression of several mRNAs. Therefore, we established a CNC network to deeply analyze the connections between dysregulated lncRNAs and mRNAs. Some major linked mRNAs have been described as being connected to DLE, including lnc-DDX47-3. We thus supposed that lncRNAs may be related to the pathogenesis of DLE via a regulation of gene co-expression (IL19, CXCL1, CXCL11, and TNFSF15).
Finally, we constructed another network of lncRNA-TFs. Emerging evidence has shown that some TFs are involved in the progression of DLE. Therefore, a compositive analysis of TFs and differential co-expression genes may help to better understand the pathogenesis of DLE. In this study, we found that STAT4, ETV6, and ZNF597 were the most important TFs. Several studies identified a few of them as possible factors promoting the progression of DLE, but, the underlying mechanisms for this process remain elusive. Several STAT4 genes targets are reported to be enriched to functional pathways in the type I interferon system, and to have key roles in inflammatory response (34-36). Therefore, STAT4 has ability to influence the regulation many target genes, which may be closely related to their relationship with DLE. However, further study is needed to investigate the relationship between lncRNA-TFs and DLE.
In summary, we underlined a profile of dysregulated mRNAs/lncRNAs/circRNAs which may be prospectively useful in clinical practice for identifying possible markers with critical roles in the development of DLE. These results have revealed that certain mRNAs/lncRNAs/circRNAs may have substantial value in DLE diagnosis and therapy.
Acknowledgments
Funding: The present study was supported by the National Natural Science Foundation of China (grant no: 81271157), National Construction Project of Clinical Key Specialized Department [(2013)544].
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, and the questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The local ethics committee of Shanghai Ninth People’s Hospital approved this study.
References
- Ghauri AJ, Valenzuela AA, O'Donnell B, et al. Periorbital discoid lupus erythematosus. Ophthalmology 2012;119:2193-2194.e11. [Crossref] [PubMed]
- Kuhn A, Landmann A. The classification and diagnosis of cutaneous lupus erythematosus. J Autoimmun 2014;48-49:14-9. [Crossref] [PubMed]
- Méndez-Flores S, Hernández-Molina G, Azamar-Llamas D, et al. Inflammatory chemokine profiles and their correlations with effector CD4 T cell and regulatory cell subpopulations in cutaneous lupus erythematosus. Cytokine 2019;119:95-112. [Crossref] [PubMed]
- Solé C, Domingo S, Ferrer B, et al. MicroRNA Expression Profiling Identifies miR-31 and miR-485-3p as Regulators in the Pathogenesis of Discoid Cutaneous Lupus. J Invest Dermatol 2019;139:51-61. [Crossref] [PubMed]
- Tenti S, Fabbroni M, Mancini V, et al. Intravenous Immunoglobulins as a new opportunity to treat discoid lupus erythematosus: A case report and review of the literature. Autoimmun Rev 2018;17:791-5. [Crossref] [PubMed]
- Moirangthem A, Patel T. Circular RNA: non-coding RNA with emerging roles in stem cell differentiation. Transl Cancer Res 2018;7:S50-2. [Crossref]
- Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev 2016;96:1297-325. [Crossref] [PubMed]
- Dragomir M, Chen B, Calin GA. Exosomal lncRNAs as new players in cell-to-cell communication. Transl Cancer Res 2018;7:S243-52. [Crossref] [PubMed]
- Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol 2017;35:177-98. [Crossref] [PubMed]
- Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol 2017;18:962-72. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nature reviews. Genetics 2016;17:47-62. [Crossref] [PubMed]
- Diamantopoulos MA, Tsiakanikas P, Scorilas A. Non-coding RNAs: the riddle of the transcriptome and their perspectives in cancer. Ann Transl Med 2018;6:241. [Crossref] [PubMed]
- Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nature reviews. Nat Rev Mol Cell Biol 2018;19:143-57. [Crossref] [PubMed]
- Uszczynska-Ratajczak B, Lagarde J, Frankish A, et al. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet 2018;19:535-48. [Crossref] [PubMed]
- Flemming A. The enigma of circular RNA. Nat Rev Immunol 2019;19:351. [Crossref] [PubMed]
- Litke JL, Jaffrey SR. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat Biotechnol 2019;37:667-75. [Crossref] [PubMed]
- Wilusz JE. Circle the Wagons: Circular RNAs Control Innate Immunity. Cell 2019;177:797-9. [Crossref] [PubMed]
- Vo JN, Cieslik M, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell 2019;176:869-881.e13. [Crossref] [PubMed]
- Liu CX, Li X, Nan F, et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019;177:865-880.e21. [Crossref] [PubMed]
- Zhu P, Zhu X, Wu J, et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat Immunol 2019;20:183-94. [Crossref] [PubMed]
- Zlotorynski E. The innate function of circular RNAs. Nature reviews. Nat Rev Mol Cell Biol 2019;20:387. [Crossref] [PubMed]
- Kopp W, Vingron M. An improved compound Poisson model for the number of motif hits in DNA sequences. Bioinformatics 2017;33:3929-37. [Crossref] [PubMed]
- Li Q, Wang Y, Wu S, et al. CircACC1 Regulates Assembly and Activation of AMPK Complex under Metabolic Stress. Cell Metab 2019;30:157-173.e7. [Crossref] [PubMed]
- Abd-Elmawla MA, Fawzy MW, Rizk SM, et al. Role of long non-coding RNAs expression (ANRIL, NOS3-AS, and APOA1-AS) in development of atherosclerosis in Egyptian systemic lupus erythematosus patients. Clin Rheumatol 2018;37:3319-28. [Crossref] [PubMed]
- Li LJ, Zhao W, Tao SS, et al. Comprehensive long non-coding RNA expression profiling reveals their potential roles in systemic lupus erythematosus. Cell Immunol 2017;319:17-27. [Crossref] [PubMed]
- Wang Y, Chen S, Chen S, et al. Long noncoding RNA expression profile and association with SLEDAI score in monocyte-derived dendritic cells from patients with systematic lupus erythematosus. Arthritis Res Ther 2018;20:138. [Crossref] [PubMed]
- Ye H, Wang X, Wang L, et al. Full high-throughput sequencing analysis of differences in expression profiles of long noncoding RNAs and their mechanisms of action in systemic lupus erythematosus. Arthritis Res Ther 2019;21:70. [Crossref] [PubMed]
- Cortes R, Forner MJ. Circular RNAS: novel biomarkers of disease activity in systemic lupus erythematosus? Clin Sci (Lond) 2019;133:1049-52. [Crossref] [PubMed]
- Li S, Zhang J, Tan X, et al. Microarray expression profile of circular RNAs and mRNAs in children with systemic lupus erythematosus. Clin Rheumatol 2019;38:1339-50. [Crossref] [PubMed]
- Miao Q, Zhong Z, Jiang Z, et al. RNA-seq of circular RNAs identified circPTPN22 as a potential new activity indicator in systemic lupus erythematosus. Lupus 2019;28:520-8. [Crossref] [PubMed]
- Wang X, Zhang C, Wu Z, et al. CircIBTK inhibits DNA demethylation and activation of AKT signaling pathway via miR-29b in peripheral blood mononuclear cells in systemic lupus erythematosus. Arthritis Res Ther 2018;20:118. [Crossref] [PubMed]
- Ayoub S, Hickey MJ, Morand EF. Mechanisms of disease: macrophage migration inhibitory factor in SLE, RA and atherosclerosis. Nat Clin Pract Rheumatol 2008;4:98-105. [Crossref] [PubMed]
- Santos LL, Morand EF. Macrophage migration inhibitory factor: a key cytokine in RA, SLE and atherosclerosis. Clin Chim Acta 2009;399:1-7. [Crossref] [PubMed]
- Ciccacci C, Perricone C, Ceccarelli F, et al. A multilocus genetic study in a cohort of Italian SLE patients confirms the association with STAT4 gene and describes a new association with HCP5 gene. PloS One 2014;9:e111991. [Crossref] [PubMed]
- Korman BD, Kastner DL, Gregersen PK. STAT4: genetics, mechanisms, and implications for autoimmunity. Curr Allergy Asthma Rep 2008;8:398-403. [Crossref] [PubMed]
- Wang C, Sandling JK, Hagberg N, et al. Genome-wide profiling of target genes for the systemic lupus erythematosus-associated transcription factors IRF5 and STAT4. Ann Rheum Dis 2013;72:96-103. [Crossref] [PubMed]


