
miR-1245a promotes the proliferation and invasion of colon adenocarcinoma by targeting BRCA2
Introduction
Colon cancer is the fourth leading cause of cancer-related deaths (1). Among all histological types, colon adenocarcinoma (CA) is the most common one (2). Recently, the treatment for CA, including surgery, radiation therapy, chemotherapy, and targeted therapy, has been improved; however, the prognosis of CA is unsatisfactory. The blood test markers, such as CEA and CA199, lack good specificity and sensitivity for screening the lesions for all patients. Therefore, it is important to further understand the molecular mechanism of CA progression and to identify the desired prognosis biomarkers of CA.
MicroRNA (miRNA) is a small noncoding RNAs with 18–25 nucleotides (3). It has reported that mature miRNAs could bind to the 3'UTR of target mRNAs, regulating the downstream genes (4). Increasing studies have reported that miRNA was associated with prognosis and diagnosis for various cancers, and miRNAs might play an important role in the development of CA (5-9). For example, Zheng et al. reported that miR-1307 inhibits tumor progression by targeting ISM1 in colon cancer (10). Yu et al. indicated that miR-21-5p promotes the progression of CA cells by targeting CHL1 (11). Therefore, miRNA might be a promising biomarker for CA.
MicroRNA-1245 (miR-1245) exerts its tumorigenesis effect in tumor progression because a previous study demonstrated that it regulates the tumor suppressor protein BRCA2 negatively (12). In addition, it was reported that the expression of NKG2D, an activating receptor involved in tumor immunosurveillance, was inhibited by miR-1245 in natural killer cells (13). In breast cancer, miR-1245 targeted BRCA2 directly and suppressed its translation (14). Recently, Yang et al. indicated that miR-1245 promotes the progression of lung cancer cells by targeting BRCA2 (15). However, how miR-1245a, a member of the miR-1245 family, influences the progression of CA remains unknown. Therefore, further study for miR-1245a and CA is needed.
In this study, the function of miR-1245a in CA cells was explored through CCK8 assays, colony formation assays, and Transwell assays. Then we investigated the relationship between miR-1245a expression level and the prognosis of CA patients using the TCGA database. Furthermore, the miR-1245a/BRACA2 axis on the CA was detected in vitro. These findings implied that miR-1245a might be a potential therapeutic target for CA.
Methods
In the present study, the counts of CA miR-1245a expression profiles were acquired from the Cancer Genome Atlas (TCGA) database in September 2019. The eligibility criteria used to screen the included patients were as follows: (I) patients with certain TNM stage; (II) histologically confirmed CA; (III) overall survival (OS) time was no less than 3 months. For the eligible patients, clinical characteristics including age, T stage, N stage, TNM stage were collected. The CA patients were classified into a low-risk group and a high-risk-group with the median of miR-1245a expression.
Ethical statement and tissue collection
The Ethics Committee approved this study of the Sixth Affiliated Hospital of Sun Yat-sen University. The CA tissues and adjacent normal samples were collected from patients who had undergone surgery recently. These 20 pairs of tumor and adjacent tissue specimens were frozen immediately and stored at −80 °C. Informed consent from all patients was obtained in this research.
Total RNA extraction and qRT-PCR
We extracted total RNA with RNAiso Plus (TaKaRa, Japan). The purity and concentration of the total RNA samples were measured with a NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA). All cDNA was generated with PrimeScript RT reagent Fix (TaKaRa, Japan). qRT-PCR for miRNAs, and the U6 was performed on a LightCycler® 96 System (Roche, Switzerland).
Cell culture
CA cells (DLD-1, SW480, LoVo, and T84 cell lines) and normal colonic epithelial cells (NCM-460 cell line) were purchased from the Shanghai Institutes for Biological Science, China. Colonic cells were cultured in DMEM (Gibco; Thermo Fishier Scientific, Suzhou, China) with 10% fetal bovine plasma at 37 °C in a humidified atmosphere with 5% CO2.
Transfection
Before transfection, CA cells were seeded in 6-well plates to 60% confluence. miRNA mimics and inhibitors, BRCA2 siRNA and pcDNA3.1 BRCA2, were transfected using a Lipidosome2000 transfection kit (Invitrogen, USA) following the manufacturer’s instructions.
Cell proliferation assay
CCK8 assays were performed to explore the effect of miR-1245a in CA cell proliferation. CA cells transfected with miRNA mimics or inhibitors were seeded into 96-well plates at 5×103/well and cultured for 24, 48, 72, 96, 120 and 144 h. Cell proliferation was determined using a Cell Counting Kit-8 (CCK-8; ImmunoWay Biotechnology Company Plano, TX, USA), and OD450 values were monitored. Colony-forming assays were used to evaluate the clonogenic ability of transfected CA cells. CA cells were seeded into 6-well plates at a density of 1,000 per well. After incubation for 10 days, the visible colonies were counted after staining with crystal violet.
Transwell invasion assays
The invasion ability of CA cells was evaluated by Transwell assays using Transwell chambers (Costar, USA) precoated with Matrigel. Briefly, after transfection, 2×105 CA cells in serum-free medium were added to the upper chambers (pore size, 8 µm; Corning Inc., Tewksbury, MA, USA). DMEM with 10% fetal bovine serum was added to the bottom well. After 24 h of incubation, the CA cells migrated into the lower chambers were fixed in 4% paraformaldehyde. Subsequently, the membrane was stained with a crystal violet staining solution. We counted and photographed under 100× magnification.
Luciferase reporter assay
BRCA2 sequences containing wild-type or mutated miR-1245a binding sites were synthesized and inserted into luciferase vectors, respectively. Then we seeded SW480 cells into 24-well plates (3×104 cells/well). After 48 h, miR-1245a mimics and constructed luciferase plasmids were co-transfected into SW480 cells. Finally, we used a dual-luciferase reporter assay system (Promega, USA) to measure luciferase activity.
Western blot analysis
We extracted protein using RIPA buffer (CWBIO, China). The proteins were electrophoresed by SDS-PAGE and were transferred to nitrocellulose membranes. Then proteins were incubated with primary antibodies specific for BRCA2 (diluted 1:1,000, Proteintech, USA) and GAPDH (diluted 1:1,000, ABclonal, China) at 4 °C overnight. Subsequently, the membranes were incubated with secondary antibodies. Finally, we detected signals with images acquisition using the Immobilon ECL substrate (Millipore, Germany) and Optimax X-ray Film Processor (Protec, Germany).
Statistical analysis
The relationship between miR-1245a expression and clinical characteristics was analyzed by the Chi-square test. OS was assessed by a Kaplan-Meier analysis and compared by a log-rank test. We used univariate and multivariate Cox proportional hazards regression model to analyze the independent prognosis factors. P<0.05 was considered statistically significant. Data analyses were performed using PRISM (GraphPad Software Inc., San Diego, CA, USA) and Stata version 13.1 (StataCorp., College Station, TX, USA).
Results
miR-1245a is upregulated in CA and related to poor survival
We examined miR-1245a expression in 20 pairs of CA tissues and adjacent normal tissues and found that miR-1245a was significantly higher in cancer tissues than in normal tissues (Figure 1A). In addition, the expression of miR-1245a was up-regulated in four kinds of CA cell lines, especially in SW480 cells compared with NCM-460 cells (Figure 1B). Therefore, the SW480 cell line was chosen for further experiments.
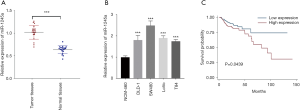
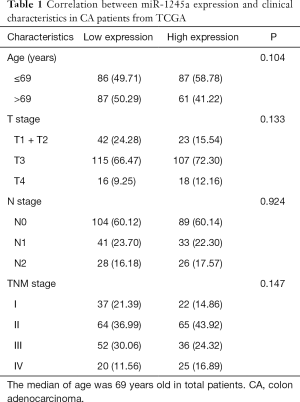
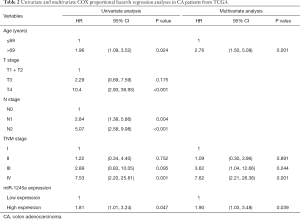
Furthermore, we analyzed the clinical relevance of miR-1245a expression in CA patients from TCGA after patients were classified into high and low-level groups via the median miR-1245a expression (Table 1). Briefly, it seems that miR-1245a had no close relationship with these clinical features, including age, T stage, N stage, and TNM stage. Kaplan-Meier survival analyze indicated that higher expression of miR-1245a was significantly associated with poor OS in patients with CA (P=0.0439) (Figure 1C). Finally, univariate and multivariate COX analyses were performed (Table 2). We found that the expression of miR-1245a was an independent prognostic factor for CA prognosis (HR =1.90; 95% CI: 1.03–3.48; P=0.039).
Full table
Full table
miR-1245a promotes the proliferation and invasion of CA cells
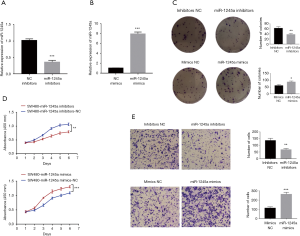
After transfection with miRNA inhibitors, miR-1245a expression in SW480 cells was down-regulated (Figure 2A). In addition, miR-1245a expression was up-regulated, followed by transfected with miR-1245a mimics (Figure 2B). Colony-formation assays illustrated that the clonogenic ability of CA cells was suppressed by miR-1245a downregulation. CCK8 assays were performed to investigate the functions of miR-1245a in cellular proliferation. Compared to the negative control conditions, down-regulated miR-1245a can suppress CA cell proliferation. Then, according to the results of Transwell assays, miR-1245a downregulation inhibited CA cell invasion abilities. In contrast, the reverse effects were yielded by miR-1245a upregulation. Overexpression of miR-1245a in CA cells promoted the proliferation and increased the invasion abilities in CA cells (Figure 2C,D,E).
miR-1245a inhibited the expression of BRCA2
Using TargetScan, the target genes of miR-1245a were predicted, and we found that a binding site existed in the 3'UTR region of BRCA2 (Figure 3A). Since the regulatory effect of miR-1245 on BRCA2 expression have reported in previous studies as BRCA2 was a target gene of miR-1245, we speculated that miR-1245a might promote CA development via inhibiting BRCA2. Furthermore, we found that the luciferase reporter activity was decreased in only cells co-transfected with miR-1245a mimics and BRCA2-wild type containing the complete miR-1245a sequence, and overexpression of miR-1245a did not affect the luciferase activity of the vector with the mutant miR-1245a binding site in cells (Figure 3B). In addition, BRCA2 expression was inhibited by the up-regulated miR-1245a, while miR-1245a downregulation increased BRCA2 expression (Figure 3C). Therefore, miR-1245a may inhibit BRCA2 expression by binding the 3'UTR of BRCA2 in CA cells.
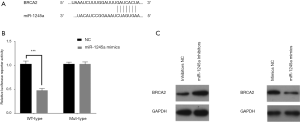
BRCA2 inhibits the progression of CA cells
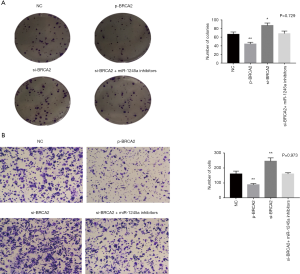
After transfection, colony-formation assays illustrated that the SW480 cell viability was suppressed by the overexpression of BRCA2 while it was enhanced by the silencing of BRCA2. Additionally, the effect of BRCA2 silence was restored by miR-1245a inhibitors (Figure 4A). In Transwell assay, it indicated that the invasion ability of SW480 cells was inhibited by BRCA2 overexpression plasmid. On the other hand, cell invasion ability was enhanced in the CA cells transfected with si-BRCA2, and it was inhibited by miR-1245a inhibitors (Figure 4B). In brief, BRCA2 might inhibit CA progression as proliferation and invasion.
Discussion
In the present study, we explored the relationship between the miR-1245a and the prognosis of CA patients from TCGA data as well as miR-1245a expression in CA tissues and CA cells. The results of functional assays indicated that miR-1245a promotes the proliferation and invasion ability of CA cells. Additionally, we also found that miR-1245a inhibited the expression of BRCA2. According to the results of the rescue experiment, it suggested that miR-1245a promotes CA cell proliferation and invasion by inhibiting BRCA2.
As a common type of colon cancer, CA is malignant cancer with high incidence worldwide. In current, the therapeutic strategies and the screen way for CA have been improved. However, CA patients with advance stages have poor survival due to the development of chemoresistance (16). Dysregulated miRNAs play a key role in the progression of various cancers, suggesting it could be a promising biomarker. As previously reported, miR-1245 expression was up-regulated in cancer tissues, and these studies indicated that miR-1245 promotes cancer progression in breast cancer and lung cancer (14,15). To further investigate the role of miR-1245a in colon cancer, a series experiments were performed. These results reveal that miR-1245a downregulation inhibits the development in vitro, which further identified its tumorigenesis in CA.
BRCA2 is a susceptibility gene, and its mutation was frequently found in cancer (17,18). According to the previous study, down-regulated BRCA2 was found in lung cancer tissues and implied that it was a tumor-suppressing gene (15). Similarly, the protein expression of BRCA2 is often decreased in sporadic breast cancer (14). To investigate the regulatory relationship between BRCA2 and miR-1245a in CA, bioinformatics and luciferase reporter assay were performed. We found that miR-1245a could bind directly to the 3'UTR of BRCA2. The CA cell invasion induced by the silencing of BRCA2 could be inhibited by miR-1245a down-regulation, which further suggested that BRCA2 might be a putative tumor suppressor gene in CA. There were only a few studies about the connection between BRCA2 and the development of colon cancer. For example, a study reported that BRCA2 on chromosome 13 demonstrated a high frequency of allelic imbalance in primary colon carcinoma, which suggests an involvement of BRCA2 in colon tumorigenesis (19). Besides, Luca et al. reported that BRCA2 deletion induces alternative lengthening of telomeres in positive telomerase colon cancer cells (20). In present study, we explore the relationship between miR-1245a and BRCA2 in CA, which might suggest a new molecular mechanism for colon cancer. However, more studies are needed to confirm the precise role and the prognostic significance of BRCA2 in colon cancer.
Although some meaningful results were observed, several limitations exist in our study. First, we explored the relationship between miR-1245a expression and CA prognosis using the TCGA database; however, real-world studies of multicenter should be further performed to investigate the role of miR-1245a. Second, we only explore the effect of miR-1245a in vitro; therefore, further animal studies were needed. Third, we focus on the functions of miR-1245a in promoting proliferation and invasion of CA cells; however, the complete understanding of miR-1245a in CA cells remains unclear, suggesting that detail underlying mechanism should be explored in the development of CA.
Conclusions
In conclusion, miR-1245a was up-regulated in CA cells and tissues. Additionally, increased miR-1245a was associated with poor survival in CA patients from the TCGA database. Furthermore, our data indicated that up-regulated miR-1245a promotes cancer progression. Our results suggested that miR-1245a could be a potential biomarker for CA progression.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China [grant numbers 81973858], the China Postdoctoral Science Foundation (2019M653197).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Ethics Committee approved this study of the Sixth Affiliated Hospital of Sun Yat-sen University. Informed consent from all patients was obtained in this research.
References
- Cassidy S, Syed BA. Colorectal cancer drugs market. Nat Rev Drug Discov 2017;16:525-6. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 2002;30:363-4. [Crossref] [PubMed]
- Mondaca S, Yaeger R. Colorectal cancer genomics and designing rational trials. Ann Transl Med 2018;6:159. [Crossref] [PubMed]
- Su C, Huang DP, Liu JW, et al. miR-27a-3p regulates proliferation and apoptosis of colon cancer cells by potentially targeting BTG1. Oncol Lett 2019;18:2825-34. [PubMed]
- Lin Q, Ma L, Wang D, et al. A novel Camptothecin analogue inhibits colon cancer development and downregulates the expression of miR-155 in vivo and in vitro. Transl Cancer Res 2017;6:511-20. [Crossref]
- Chai B, Guo Y, Cui X, et al. MiR-223-3p promotes the proliferation, invasion and migration of colon cancer cells by negative regulating PRDM1. Am J Transl Res 2019;11:4516-23. [PubMed]
- Shao Q, Xu J, Deng R, et al. SNHG 6 promotes the progression of Colon and Rectal adenocarcinoma via miR-101-3p and Wnt/beta-catenin signaling pathway. BMC Gastroenterol 2019;19:163. [Crossref] [PubMed]
- Zheng Y, Zheng Y, Lei W, et al. miR-1307-3p overexpression inhibits cell proliferation and promotes cell apoptosis by targeting ISM1 in colon cancer. Mol Cell Probes 2019.101445. [Epub ahead of print]. [Crossref] [PubMed]
- Yu W, Zhu K, Wang Y, et al. Overexpression of miR-21-5p promotes proliferation and invasion of colon adenocarcinoma cells through targeting CHL1. Mol Med 2018;24:36. [Crossref] [PubMed]
- Arbini AA, Guerra F, Greco M, et al. Mitochondrial DNA depletion sensitizes cancer cells to PARP inhibitors by translational and post-translational repression of BRCA2. Oncogenesis 2013;2:e82. [Crossref] [PubMed]
- Espinoza JL, Takami A, Yoshioka K, et al. Human microRNA-1245 down-regulates the NKG2D receptor in natural killer cells and impairs NKG2D-mediated functions. Haematologica 2012;97:1295-303. [Crossref] [PubMed]
- Song L, Dai T, Xie Y, et al. Up-regulation of miR-1245 by c-myc targets BRCA2 and impairs DNA repair. J Mol Cell Biol 2012;4:108-17. [Crossref] [PubMed]
- Yang L, Wang J, Fan Y, et al. Hsa_circ_0046264 up-regulated BRCA2 to suppress lung cancer through targeting hsa-miR-1245. Respir Res 2018;19:115. [Crossref] [PubMed]
- Germani A, Matrone A, Grossi V, et al. Targeted therapy against chemoresistant colorectal cancers: Inhibition of p38alpha modulates the effect of cisplatin in vitro and in vivo through the tumor suppressor FoxO3A. Cancer Lett 2014;344:110-8. [Crossref] [PubMed]
- Zhang J, Sun J, Chen J, et al. Comprehensive analysis of BRCA1 and BRCA2 germline mutations in a large cohort of 5931 Chinese women with breast cancer. Breast Cancer Res Treat 2016;158:455-62. [Crossref] [PubMed]
- Sugano K, Nakamura S, Ando J, et al. Cross-sectional analysis of germline BRCA1 and BRCA2 mutations in Japanese patients suspected to have hereditary breast/ovarian cancer. Cancer Sci 2008;99:1967-76. [Crossref] [PubMed]
- Sivarajasingham NS, Cawkwell L, Baker RP, et al. Implication of the BRCA2 and putative "BRCA3" genes in Dukes' stage C, replication error-negative colon cancer. Ann Surg Oncol 2006;13:881-6. [Crossref] [PubMed]
- Pompili L, Maresca C, Dello Stritto A, et al. BRCA2 Deletion Induces Alternative Lengthening of Telomeres in Telomerase Positive Colon Cancer Cells. Genes (Basel) 2019. [Crossref] [PubMed]


