
Automated quantitative assessment of oncological disease progression using deep learning
Assessing oncological disease progression is a major part of radiologists’ work. Today, such assessment is usually done by either eyeballing or few 2D diameter measurements.
A common protocol for assessing disease progression is The Response Evaluation Criteria in Solid Tumours (RECIST), updated in 2009 (RECIST 1.1). It includes instructions about the use of diameter measurements for evaluation of tumour burden. The RECIST criteria have gained widespread adoption and are widely used particularly in oncology clinical trials (1-3). The RECIST 1.1 requires one diameter measurements of up-to 5 target lesions total, up-to 2 lesions per organ. It also includes the assessment of non-target lesions, basically eyeballing of disease progression.
In neuro-oncology, disease progression is assessed using Response Assessment in Neuro-Oncology (RANO) criteria (4,5). The RANO criteria are widely used in clinical trials (6). They are also increasingly used in routine clinical practice (7). These criteria rely mainly on the assessment of disease progression by MRI. They enable qualitative and quantitative evaluation of tumour burden before, during, and after therapy. In RANO, a measurable tumour is measured using two diameters. These include the maximal diameter of the tumour and a second perpendicular diameter.
RECIST, RANO and other radiology protocols operate under similar assumptions. These assumptions are that few 2D diameters can accurately represent tumour burden. Moreover, in RECIST, the summation of several diameters from different lesions aims to represent the whole-body tumour burden. These assumptions come from necessity. Providing a few 2D diameter measurements can be done quickly by radiologists. However, whole-body volumetric measurements of tumour burden are not humanly feasible. Several factors make precise manual volumetric measurements un-feasible. Firstly, the number of oncological scans is rising. Secondly, lesions can appear in complex 3D shapes. And lastly, oncological studies can have numerous metastases. Several previous works compared 2D measurements and volumetric measurements. These works showed that volumetric measurements are more reliable and accurate than 2D measurements (8-10).
A newly published paper in Lancet Oncology by Dr. Philipp Kickingereder et al., addresses automatic brain lesions quantification (11). Their work, “Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study”, presents a deep learning model for automatic quantification of brain lesions.
Deep learning is a family of algorithms which made a major impact on industry, academia, and public attention in the last few years. These algorithms are based on artificial neural networks (ANNs). Deep learning is considered a subclass of artificial intelligence (AI) (12,13).
A major focus of deep learning is the analysis of images, which is termed computer vision. Convolutional neural networks (CNNs) are the main deep learning algorithms used for computer vision. CNN are a type of ANN, aimed at recognizing repeating patterns. The hypothesis in the basis of CNN is that images contain multiple repeating patterns.
CNN were developed in the 1980s and 1990s (14,15). Yet, CNN made a major impact on the world in 2012. In that year, a CNN algorithm won the 2012 ImageNet challenge (16). ImageNet is a repository of millions of labeled images (17). These images are of world objects such as animals, humans and animate objects. In the ImageNet challenge, algorithms try to classify these millions of images into their respective 1,000 labels. In 2012, Krizhevsky and Hinton et al. won the ImageNet challenge using a CNN that showed remarkable abilities in image classification (16). An article in the Economist stated that, “Suddenly people started to pay attention, not just within the AI community but across the technology industry as a whole.” (18).
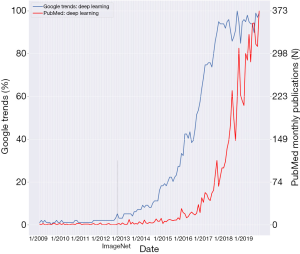
Figure 1 shows Google Trends and PubMed searches for the term “Deep Learning”. Data is from the timeframe 1/2009–11/2019. The date of the ImageNet 2012 challenge is marked on the graph. Google searches for “Deep Learning” started an exponential rise at the end of 2012. This rise reached a plateau around 2017. PubMed “Deep Learning” publications began to rise exponentially at the end of 2015. This signifies a lag of about three years from the rise in Google search.
Since 2012, CNN algorithms continue to improve, reaching human level in several image analysis tasks. A more recent deep learning advancement are generative adversarial networks (GANs) (19). These networks are a type of deep learning models aimed at generating “fake” realistically looking images. GAN are now at the center of public attention due to “deepfake” digital media manipulations. “deepfake” uses GAN to generate “fake” images of humans (20). GAN has recently been introduced into the medical field. For example, GAN have been used to transfer between imaging modalities (21,22). A work by Ben-Cohen et al. utilized GAN to produce “virtual PET” images from CT images (21).
Another important application of deep learning from recent years is natural language processing (NLP). NLP enables the conversion of human language into structured data. Similarly to images, text contains repeating patterns. This is why CNN has been successfully utilized for text analysis. In 2018, innovations in deep learning NLP led to the claim “NLP’s ImageNet moment has arrived” (23). Deep learning for NLP is increasingly being utilized in medical research (24) and industry.
The study mentioned above by Kickngereder et al. presents an example of using CNN for a computer vision task. In their work, the authors built a CNN for segmenting the borders of brain tumours. Segmentation is one of three major computer vision tasks. The other two being image classification and detection (13). In image classification, the network classifies an entire image. For example the network will classify either “image of a normal brain” vs. “image of a brain tumor”. In image detection, the network locates an object in the image, usually with a box-plot. In image segmentation, the network delineates the pixel-wise borders of an object; usually a lesion (such as a brain tumor), or an organ. The authors used the most common CNN architecture for segmentation, which is called U-Net (25).
The authors trained their CNN on MRI data from 455 patients with brain tumours. These patients were being treated at Heidelberg hospital. The authors tested the model on 2,034 MRI scans from 532 patients at 34 different institutions. Their model showed high precision. It had DICE coefficients of 0.91 for contrast-enhanced T1 weighted sequences and 0.93 for T2 weighted sequences. Moreover, the model better predicted the time to disease progression than RANO measurements. The neural network’s hazard ratio for disease progression was 2.59 compared to a hazard ratio of 2.07 for RANO. The authors concluded that CNN enabled objective and automated assessment of tumour response in neuro-oncology at high throughput. They stated that their model could serve as a blueprint for the application of CNN to improve clinical decision making (11).
As stated, the CNN was trained on 455 scans from one medical center and tested on 2,034 scans from multiple centers. When training CNN, the convention is to use a larger portion of the data for training. This is because networks improve in relation to the amount of training data. So, it will be interesting to evaluate training the CNN on the larger multi-center dataset.
A possible use of Kickngereder et al. work is the standardization of MRI disease progression assessment. Thus, the fact that the authors published their code in the open-source XNAT framework is important. Nevertheless, there is a significant gap between open-source codes and clinical use. And so, as stated by the authors, further prospective clinical evaluation is needed.
Algorithms such as Kickngereder et al. can improve patient care by performing tasks that are not humanly feasible. This should come as no surprise. Although human brains can perform trillions of calculations per second, some tasks are better done by computers; for example, algebraic calculations of very large numbers or accessing millions of lines in a spreadsheet. Automatic volumetric segmentation of tumours seems to also be such a task.
Future works should apply the present model to whole-body tumour burden quantification. This could improve the currently used RECIST criteria. Moreover, volume is not the only feature that can be extracted from radiology studies. Other features, such as intensity and texture, have been used in radiomics. Future algorithms could improve prediction of disease progression using “deep-radiomics”, utilizing CNN to learn hidden patterns in the scans. Another venue for research could be the development of networks that integrate clinical data to the imaging data. Lastly, sub-analyses should be performed on different kind of oncological diseases.
Deep learning innovations of recent years are in the progress of making a major impact on human society. In the field of oncological-radiology, they have the potential to improve patient care by facilitating precision medicine.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Schwartz LH, Litiere S, de Vries E, et al. RECIST 1.1-Update and clarification: From the RECIST committee. Eur J Cancer 2016;62:132-7. [Crossref] [PubMed]
- Nishino M, Jagannathan JP, Ramaiya NH, et al. Revised RECIST guideline version 1.1: What oncologists want to know and what radiologists need to know. AJR Am J Roentgenol 2010;195:281-9. [Crossref] [PubMed]
- van den Bent MJ, Wefel JS, Schiff D, et al. Response assessment in neuro-oncology (a report of the RANO group): assessment of outcome in trials of diffuse low-grade gliomas. Lancet Oncol 2011;12:583-93. [Crossref] [PubMed]
- Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol 2010;28:1963-72. [Crossref] [PubMed]
- Wen PY, Chang SM, Van den Bent MJ, et al. Response Assessment in Neuro-Oncology Clinical Trials. J Clin Oncol 2017;35:2439-49. [Crossref] [PubMed]
- Thust SC, Heiland S, Falini A, et al. Glioma imaging in Europe: A survey of 220 centres and recommendations for best clinical practice. Eur Radiol 2018;28:3306-17. [Crossref] [PubMed]
- Chow DS, Qi J, Guo X, et al. Semiautomated volumetric measurement on postcontrast MR imaging for analysis of recurrent and residual disease in glioblastoma multiforme. AJNR Am J Neuroradiol 2014;35:498-503. [Crossref] [PubMed]
- Sorensen AG, Patel S, Harmath C, et al. Comparison of diameter and perimeter methods for tumor volume calculation. J Clin Oncol 2001;19:551-7. [Crossref] [PubMed]
- Sorensen AG, Batchelor TT, Wen PY, et al. Response criteria for glioma. Nat Clin Pract Oncol 2008;5:634-44. [Crossref] [PubMed]
- Kickingereder P, Isensee F, Tursunova I, et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol 2019;20:728-40. [Crossref] [PubMed]
- Klang E. Deep learning and medical imaging. J Thorac Dis 2018;10:1325-8. [Crossref] [PubMed]
- Soffer S, Ben-Cohen A, Shimon O, et al. Convolutional Neural Networks for Radiologic Images: A Radiologist's Guide. Radiology 2019;290:590-606. [Crossref] [PubMed]
- LeCun Y, Bengio Y. Convolutional networks for images, speech, and time series. In: Michael AA, editor. The handbook of brain theory and neural networks. MIT Press, 1998:255-8.
- LeCun Y, Boser B, Denker JS, et al. Backpropagation applied to handwritten zip code recognition. Neural Comput 1989;1:541-51. [Crossref]
- Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Proceedings of the 25th International Conference on Neural Information Processing Systems - Volume 1; Lake Tahoe, Nevada. 2999257: Curran Associates Inc., 2012:1097-105.
- Russakovsky O, Deng J, Su H, et al. ImageNet Large Scale Visual Recognition Challenge. arXiv e-prints 2014.
- "From not working to neural networking" The Economist 25 June 2016. Retrieved 3 February 2018.
- Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2; Montreal, Canada. 2969125: MIT Press, 2014:2672-80.
- Roose K. "Here Come the Fake Videos, Too." The New York Times 2018. Available online: accessed November 2019.https://www.nytimes.com/2018/03/04/technology/fake-videos-deepfakes.html
- Ben-Cohen A, Klang E, Raskin SP, et al. Cross-Modality Synthesis from CT to PET using FCN and GAN Networks for Improved Automated Lesion Detection. arXiv e-prints 2018.
- Frid-Adar M, Klang E, Amitai M, et al. Synthetic Data Augmentation using GAN for Improved Liver Lesion Classification. arXiv e-prints 2018.
- Ruder S. NLP's ImageNet moment has arrived. Available online: https://thegradient.pub/nlp-imagenet/, 7.8.2018.
- Yuan J, Zhu H, Tahmasebi A. Classification of Pulmonary Nodular Findings based on Characterization of Change using Radiology Reports. AMIA Jt Summits Transl Sci Proc 2019;2019:285-94. [PubMed]
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv e-prints 2015.


