Low expression of A-kinase anchor protein 5 predicts poor prognosis in non-mucin producing stomach adenocarcinoma based on TCGA data
Introduction
Gastric cancer is the sixth most common malignant tumor in the world, with the third highest mortality rate (1). Although the survival rate has reached about 50% in Japan and South Korea due to great efforts made by these countries, the global survival rate of gastric cancer is still unsatisfactory (2,3). At present, the prognosis of gastric cancer is determined through clinicopathologic classification, such as TNM staging and histopathological staging. Although many patients have the same clinical and pathological stages, their prognoses are far from the same. According to the WHO histopathological classification for gastric cancer, gastric adenocarcinoma is mainly divided into tubular adenocarcinoma, papillary adenocarcinoma, mucinous adenocarcinoma, and signet ring cell carcinoma (4). Mucinous adenocarcinoma and signet ring cell carcinoma have the characteristics of being mucin-producing in its histology. They mainly belong to the undifferentiated type in the classification of gastric cancer in Japan, which generally indicates a poor prognosis (5). The prognosis of non-mucin producing stomach adenocarcinomas (NMSA) is better, but some of them still have an outcome of death. Whether or not these patients have different biological characteristics from the patients with a good prognosis is still unclear.
A-kinase anchor protein 5 (AKAP5), also named AKAP79, is a 427 amino acid protein that functions as an anchor cAMP-dependent protein kinase (PKA) to cytoskeleton or organelle-associated proteins, that transduce cAMP signals into specific intracellular effectors. AKAP5 also regulates the beta2- adrenergic receptor signaling pathway, participation in energy metabolism synthesis (6,7). Earlier studies on how AKAP5 involving disease are few. Some research has found that AKAP5 is associated with chronic obstructive pulmonary disease, long-term depression, and diabetes, but there is little research on its relationship with malignant tumors (8-10).
In this study, we mined the data of NMSA in the stomach adenocarcinoma (STAD) collection on the Cancer Genome Atlas (TCGA) database and found that AKAP5 was significantly increased in NMSA, but low AKAP5 expression of NMSA has a significantly poorer prognosis among most clinical and pathological stages. It has excellent performance for predicting the prognosis of NMSA as a biomarker. GSEA analysis was conducted to identify the biological pathway involved in AKAP5. This study suggested that the AKAP5 gene may play a key role in the pathological process of NMSA, increasing our understanding of this disease.
Methods
RNA-sequencing patient data and bioinformatics analysis
The RNA high-throughput sequencing data and corresponding clinicopathologic data of the stomach adenocarcinomas (STAD) projects were downloaded from the TCGA. The disease type of the adenomas and adenocarcinomas was included, and other types such as cystic, mucinous, and serous neoplasms were excluded. HTSeq-FPKM workflow was used for gene expression normalization. All data were conducted using the R software (version 3.5.3).
Gene set enrichment analysis (GSEA)
GSEA was utilized to understand which biological processes are involved in the high and low expression of AKAP5, respectively. In this study, we used version 4.0 of the GSEA software downloaded from the official website (11). According to the cut-off point, the samples were divided into two phenotypes: high expression and low expression. The Signal2Noise method was used to evaluate the correlation between the expression of each gene and AKAP5; then, genes were ranked according to the correlation score from high to low. The gene set permutation was run 1000 times per analysis, and the software automatically calculated the normalized enrichment score (NES), nominal P value, and false discovery rate (FDR). Gene sets with nom P value <0.05 and FDR <0.25 were significantly enriched.
Statistical analysis
All statistical analyses were performed using R (v.3.5.3). Clinical pathologic features associated with AKAP5 high and low expression groups were analyzed using the chi-square and Fisher exact tests. The relationship between overall survival (OS) and AKAP5 expression among all NMSA patients and each clinicopathological subgroup was analyzed using the Kaplan-Meier method, using the Survival package in R. The correlations between AKAP5 expression and survival along with other clinicopathological characteristic were analyzed using univariate and multivariate Cox regression analysis. The best cut point of AKAP5 expression was determined by the maximally selected rank statistics method using survminer package in R.
Results
Patient characteristics
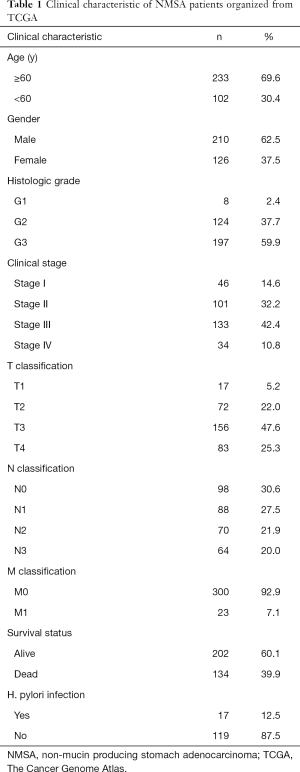
The clinical data of the 336 NMSA patients were downloaded from the TCGA database, which included patient age, gender, tumor location, histologic grade, clinical stage, TNM classification, survival status, helicobacter pylori infection (Table 1).
Full table
Differential AKAP5 expression in NMSA compared with adjacent mucosa
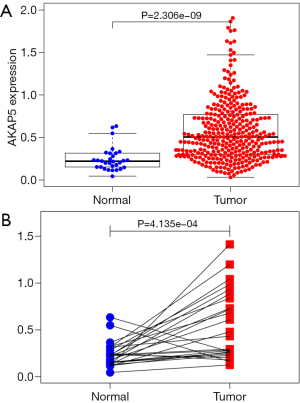
Differential expression of AKAP5 in NMSA and adjacent mucosa was performed by two independent sample Wilcoxon Rank Sum Test and paired Wilcoxon Rank Sum Test. The result showed that AKAP5 expression was increased in NMSA (all tumor vs. adjacent mucosa, P=2.306e-09, paired tumor vs. adjacent mucosa, P=4.135e-04, Figure 1).
Correlation between AKAP5 expression and clinicopathologic variables of NMSA
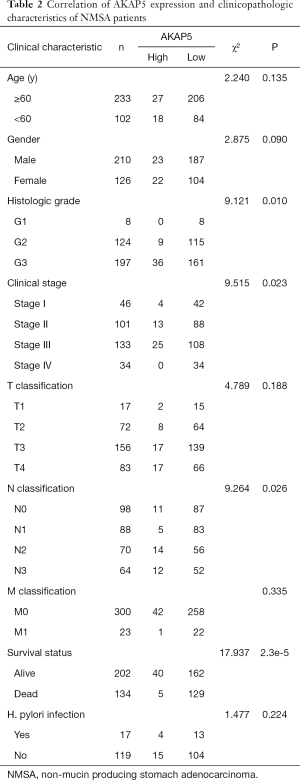
AKAP5 expression data of NMSA and their clinicopathologic information were downloaded from the TCGA. Subsequently, patients were divided into high AKAP5 expression group and low AKAP5 expression group according to the gene expression cutpoint 0.9975. The correlation between AKAP5 expression and clinicopathologic variables of NMSA is summarized in Table 2. Histologic grade, clinical stage, N classification, and survival status were significantly correlated with AKAP5 expression.
Full table
Low AKAP5 expression is a significant risk factor in predicting OS of NMSA
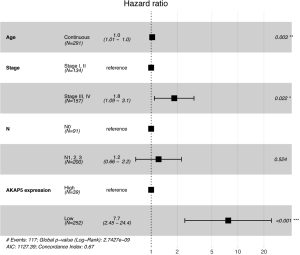
Although AKAP5 expression increased in the tumor samples, Kaplan-Meier curves showed that low AKAP5 expression was associated with a worse OS among NMSA patients (P=5.003e-05; Figure 2). Furthermore, low AKAP5 expression also indicated poor OS in clinical stage III and IV (P=4.646e-05), TNM stage T3 (P=0.016), T4 (P=0.001), N2 (P=0.012), N3 (P=0.003), M0 (P=3.911e-05), and histological grade G3 (P=1.658e-04). Univariate Cox regression analysis show that reduced AKAP5 expression in NMSA is associated with age (HR =1.03, P=0.007), stage (HR =1.84 for stage I, II vs. Stage III, IV, P=0.002) and M classification (HR =1.8 for M0 vs. M1, P=0.010). Univariate and multivariate Cox regression analysis showed that AKAP5 expression was an independent risk factor for OS among NMSA patients (HR =7.58, P=0.001). These results are described in Table 3 and Figure 3.
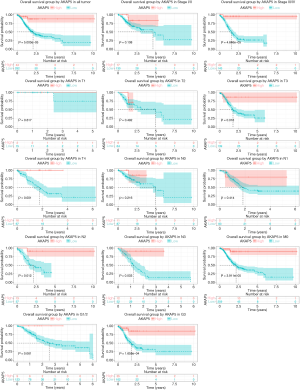
Full table
GSEA
GSEA was performed to analyze which signaling pathways were activated in the AKAP5 low and high expression groups. Data sets from MSigDB Collection (h.all.v7.0.symbols) with FDR <0.25 and nom-P value <0.05 were considered to be significantly different. The most significantly enriched signaling pathways were described in Figure 4 and Table 4. Gene sets related to cholesterol homeostasis, glycolysis, estrogen response late, adipogenesis, estrogen response early, notch signaling, and peroxisome were differentially enriched with the low AKAP5 expression phenotype.
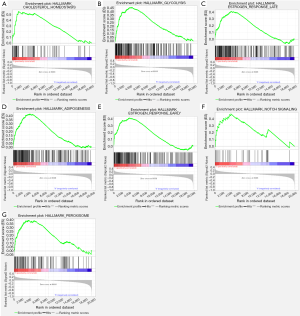
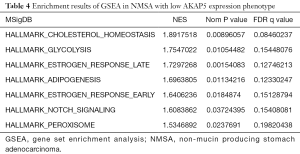
Full table
Discussion
As an essential protein family regulating the cAMP-PKA signaling pathway, AKAPs are directly related to the occurrence and development of carcinoma. For example, the specific polymorphism of AKAP9 is associated with the risk of breast cancer and has been found to be significantly higher in human colorectal cancer than in the adjacent tissues, as well as promoting cell proliferation, invasion, and migration (12). Mutations in the AKAP10 gene increase the risk of breast and colorectal cancer (13,14). AKAP12 expression is associated with endothelial barrier function and is speculated to have a tumor suppressor effect (15,16).
The AKAP protein family is an important regulatory protein of the cAMP-PKA signaling pathway, which is capable of anchoring the protein kinase A (PKA) regulatory subunits RI and RII to specific organelles specifically; thus, it is able to transduce the second messenger cAMP signal. Studies have shown that the cAMP-PKA pathway is closely related to the proliferation, apoptosis, and tumorigenesis of various tumor cells (such as prostate cancer, colorectal cancer, and breast cancer) (12,17-19).
The relationship between AKAP5 and the development of tumors is poorly understood; some studies have suggested there to be an indirect relationship with the development of them. AKAP5 can bind to E-cadherin and β-catenin to regulate mucosal adhesion junctions, which may be associated with tumor migration and metastasis (20). AKAP5 inhibits cell proliferation, but in smooth muscle cells rather than tumor cells (21). In breast cancer, low AKAP5 expression is more prone to metastasis and recurrence (22).
Since the role of AKAP5 in stomach adenocarcinoma has not been clarified, GSEA analysis was used to predict the pathway associated with AKAP5 in NMSA. The results showed that the low AKAP5 expression group was associated with cholesterol homeostasis, glycolysis, estrogen response, adipogenesis, notch signaling, and peroxisome. The estrogen signaling pathway has been widely studied in breast cancer, and this pathway also contributes to the oncogenesis and the advancement of gastric cancer as well. For instance, estrogen receptor (ER) α36 is highly expressed in gastric cancer and is associated with lymph node metastasis (23). The possible mechanisms by which estrogen receptors promote tumor growth include the activation of the Akt-PI3K signaling pathway by glucose-regulated protein 94 (GRP94), or promoting gastric cancer cell proliferation, differentiation, and invasion by activating the c-Src signaling pathway and increasing cyclins D1 expression, which regulates the cell cycle to promote proliferation (23-25). The activation of the Notch signaling pathway is related to the clinical progress of gastric cancer (26,27). It may promote the proliferation, migration, and invasion of gastric cancer cells by interacting with mTOR, STAT3-Twist, and other signaling pathways or inhibiting the activity of PTEN (27-29). Enhanced glycolysis is part of the Warburg effect of tumor cells, which enables gastric cancer cells to metabolize glucose into lactic acid in an aerobic environment, providing a source for cellular biosynthesis and cell division (30). This process may involve changes in key enzymes of glycolysis and mitochondrial damage, which are often related to a poor prognosis of the tumor (31-34). In addition to abnormal glucose metabolism, abnormal lipid metabolism is also observed in gastric cancer, but the specific mechanisms for this are still unclear (35). The research on the relationship between these pathways and AKAP5 is very scarce and needs further explored through experiments.
Unfortunately, the major deficiency of this study is that the research data is only from TCGA. Although the source of TCGA samples may have some limitations, TCGA is one of the best cancer databases in the world, with the largest number of sequencing data and the most comprehensive clinical information. In a limited sample, we found that low expression of AKAP5 may be a potential molecular marker for predicting poor prognosis of NMSA. Besides, cholesterol homeostasis, glycolysis, estrogen response, adipogenesis, notch signaling, and peroxisome may be the key pathways regulated by AKAP5 in NMSA. It also suggested that AKAP5 might have a potential biological function in stomach adenocarcinoma.
Acknowledgments
Funding: The study was funded by National Natural Science Foundation of China (No. 81373564, and No. 81302881).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Yang Y, Xing P, Zhou N, et al. The necessity of adjuvant radiotherapy for locally advanced gastric cancer in China. Transl Cancer Res 2019;8:676-82. [Crossref]
- Zhang CD, Yamashita H, Seto Y. Gastric cancer surgery: historical background and perspective in Western countries versus Japan. Ann Transl Med 2019;7:493. [Crossref] [PubMed]
- Verdecchia A, Francisci S, Brenner H, et al. Recent cancer survival in Europe: a 2000-02 period analysis of EUROCARE-4 data. Lancet Oncol 2007;8:784-96. [Crossref] [PubMed]
- Solanki S, Chakinala RC, Haq KF, et al. Inpatient burden of gastric cancer in the United States. Ann Transl Med 2019;7:772. [Crossref] [PubMed]
- Japanese Gastric Cancer Association. Japanese gastric cancer treatment guidelines 2014 (ver. 4). Gastric Cancer 2017;20:1-19. [Crossref] [PubMed]
- Robertson HR, Gibson ES, Benke TA, et al. Regulation of postsynaptic structure and function by an A-kinase anchoring protein-membrane-associated guanylate kinase scaffolding complex. J Neurosci 2009;29:7929-43. [Crossref] [PubMed]
- Gold MG, Stengel F, Nygren PJ, et al. Architecture and dynamics of an A-kinase anchoring protein 79 (AKAP79) signaling complex. Proc Natl Acad Sci U S A 2011;108:6426-31. [Crossref] [PubMed]
- Poppinga WJ, Heijink IH, Holtzer LJ, et al. A-kinase-anchoring proteins coordinate inflammatory responses to cigarette smoke in airway smooth muscle. Am J Physiol Lung Cell Mol Physiol 2015;308:L766-75. [Crossref] [PubMed]
- Woolfrey KM, O'Leary H, Goodell DJ, et al. CaMKII regulates the depalmitoylation and synaptic removal of the scaffold protein AKAP79/150 to mediate structural long-term depression. J Biol Chem 2018;293:1551-67. [Crossref] [PubMed]
- Hinke SA, Navedo MF, Ulman A, et al. Anchored phosphatases modulate glucose homeostasis. EMBO J 2012;31:3991-4004. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Hu ZY, Liu YP, Xie LY, et al. AKAP-9 promotes colorectal cancer development by regulating Cdc42 interacting protein 4. Biochim Biophys Acta 2016;1862:1172-81. [Crossref] [PubMed]
- Wirtenberger M, Schmutzhard J, Hemminki K, et al. The functional genetic variant Ile646Val located in the kinase binding domain of the A-kinase anchoring protein 10 is associated with familial breast cancer. Carcinogenesis 2007;28:423-6. [Crossref] [PubMed]
- Wang M, Zhang D, Wang R, et al. A-kinase anchoring proteins 10 expression in relation to 2073A/G polymorphism and tumor progression in patients with colorectal cancer. Pathol Oncol Res 2013;19:521-7. [Crossref] [PubMed]
- Radeva MY, Kugelmann D, Spindler V, et al. PKA compartmentalization via AKAP220 and AKAP12 contributes to endothelial barrier regulation. PLoS One 2014;9:e106733. [Crossref] [PubMed]
- Gelman IH. Suppression of tumor and metastasis progression through the scaffolding functions of SSeCKS/Gravin/AKAP12. Cancer Metastasis Rev 2012;31:493-500. [Crossref] [PubMed]
- Neary CL, Nesterova M, Cho YS, et al. Protein kinase A isozyme switching: eliciting differential cAMP signaling and tumor reversion. Oncogene 2004;23:8847-56. [Crossref] [PubMed]
- Carlson CC, Smithers SL, Yeh KA, et al. Protein kinase A regulatory subunits in colon cancer. Neoplasia 1999;1:373-8. [Crossref] [PubMed]
- Miller WR. Regulatory subunits of PKA and breast cancer. Ann N Y Acad Sci 2002;968:37-48. [Crossref] [PubMed]
- Gorski JA, Gomez LL, Scott JD, et al. Association of an A-Kinase-anchoring Protein Signaling Scaffold with Cadherin Adhesion Molecules in Neurons and Epithelial Cells. Mol Biol Cell 2005;16:3574-90. [Crossref] [PubMed]
- Indolfi C, Stabile E, Coppola C, et al. Membrane-bound protein kinase A inhibits smooth muscle cell proliferation in vitro and in vivo by amplifying cAMP-protein kinase A signals. Circ Res 2001;88:319-24. [Crossref] [PubMed]
- Kjällquist U, Erlandsson R, Tobin NP, et al. Exome sequencing of primary breast cancers with paired metastatic lesions reveals metastasis-enriched mutations in the A-kinase anchoring protein family (AKAPs). BMC Cancer 2018;18:174. [Crossref] [PubMed]
- Deng H, Huang X, Fan J, et al. A variant of estrogen receptor-alpha, ER-alpha36 is expressed in human gastric cancer and is highly correlated with lymph node metastasis. Oncol Rep 2010;24:171-6. [PubMed]
- Fu Z, Zhen H, Zou F, et al. Involvement of the Akt signaling pathway in ER-α36/GRP94-mediated signaling in gastric cancer. Oncol Lett 2014;8:2077-80. [Crossref] [PubMed]
- Wang X, Deng H, Zou F, et al. ER-α36-mediated gastric cancer cell proliferation via the c-Src pathway. Oncol Lett 2013;6:329-35. [Crossref] [PubMed]
- Yeh TS, Wu CW, Hsu KW, et al. The activated Notch1 signal pathway is associated with gastric cancer progression through cyclooxygenase-2. Cancer Res 2009;69:5039-48. [Crossref] [PubMed]
- Hsu KW, Hsieh RH, Huang KH, et al. Activation of the Notch1/STAT3/Twist signaling axis promotes gastric cancer progression. Carcinogenesis 2012;33:1459-67. [Crossref] [PubMed]
- Hibdon ES, Razumilava N, Keeley TM, et al. Notch and mTOR Signaling Pathways Promote Human Gastric Cancer Cell Proliferation. Neoplasia 2019;21:702-12. [Crossref] [PubMed]
- Kim SJ, Lee HW, Baek JH, et al. Activation of nuclear PTEN by inhibition of Notch signaling induces G2/M cell cycle arrest in gastric cancer. Oncogene 2016;35:251-60. [Crossref] [PubMed]
- Liu Y, Zhang Z, Wang J, et al. Metabolic reprogramming results in abnormal glycolysis in gastric cancer: a review. Onco Targets Ther 2019;12:1195-204. [Crossref] [PubMed]
- Wu J, Hu L, Wu F. Poor prognosis of hexokinase 2 overexpression in solid tumors of digestive system: a meta-analysis. Oncotarget 2017;8:32332-344. [PubMed]
- Stacpoole PW. Therapeutic Targeting of the Pyruvate Dehydrogenase Complex/Pyruvate Dehydrogenase Kinase (PDC/PDK) Axis in Cancer. J Natl Cancer Inst 2017. [Crossref] [PubMed]
- Zhang Y, Zhang X, Wang X, et al. Inhibition of LDH-A by lentivirus-mediated small interfering RNA suppresses intestinal-type gastric cancer tumorigenicity through the downregulation of Oct4. Cancer Lett 2012;321:45-54. [Crossref] [PubMed]
- Wang H, Zhou R, Sun L, et al. TOP1MT deficiency promotes GC invasion and migration via the enhancements of LDHA expression and aerobic glycolysis. Endocr Relat Cancer 2017;24:565-78. [Crossref] [PubMed]
- Enjoji M, Kohjima M, Ohtsu K, et al. Intracellular mechanisms underlying lipid accumulation (white opaque substance) in gastric epithelial neoplasms: A pilot study of expression profiles of lipid-metabolism-associated genes. J Gastroenterol Hepatol 2016;31:776-81. [Crossref] [PubMed]