
The E3 ligase malin plays a pivotal role in promoting nuclear glycogenolysis and histone acetylation
The discovery of de novo glycogen metabolism in the nucleus sheds light into the increasingly complex roles of glycogen in overall cellular metabolism and physiology. This discovery causes one to re-consider therapies targeting cytoplasmic glycogen metabolism. Our laboratory recently demonstrated that glycogen is both synthesized and degraded in the nucleus and nuclear glycogen represents a significant carbohydrate pool for acetyl Co-A production, which is necessary for histone acetylation and modulating the epigenetic landscape (1,2). We demonstrated that malin, an E3 ubiquitin ligase, ubiquitinates glycogen phosphorylase (GP) and that ubiquitinated GP translocates from the cytosol to the nucleus. Human non-small cell lung cancers (NSCLCs) downregulate malin to perturb GP ubiquitination and nuclear glycogenolysis, creating a tumorigenic epigenetic environment (1). By restoring malin expression and nuclear glycogenolysis in the nucleus of NSCLC cells, tumor proliferation was dramatically reduced in mice, demonstrating the importance of this signaling pathway in tumorigenesis (1).
Dr. Olivier Ferron presents an excellent editorial summarizing our findings and prognosticating on downstream signaling events. We would like to comment on some of his observations regarding the work. First, his concerns that manipulation of GP localization is not a specific enough target for cancer therapies, and second, that altering acetyl-CoA availability from one source, i.e. nuclear glycogen, could negatively impact epigenetic regulation and the metabolome.
Importantly, Dr. Ferron points out that anti-cancer strategies involving GP to date have focused on inhibiting GP activity rather than promoting it. The motivation behind these GP-inhibitory therapies lies in the concept that cancer cells rely on glycogen degradation to promote central carbon metabolism (3). However, it is important to note that despite multiple studies showing growth arrest by GP inhibition on cultured cell lines in vitro, the in vivo efficacies of such a strategy have not been rigorously demonstrated (4-6). In fact, a search at clinicaltrials.gov returns no ongoing cancer trials involving GP inhibitors. Indeed, our cell lines expressing short hairpin RNA to knockdown GP exhibited very modest growth delays, suggesting that larger responses seen during drug treatments could be due to off target effects. As pointed out by Ferron, inhibition of GP on a global scale likely produces numerous side effects given its central role in maintaining whole body glucose homeostasis. Alternatively, activating GP in the nucleus as a therapeutic target could restore normal bi-compartmentalized glycogen metabolism in a cell. However, additional investigations of the interplay between malin-ubiquitin-GP on both nuclear and cytoplasmic glycogen metabolism are needed to better understand and define a targeted therapy to normalize this pathway.
While malin ubiquitinates GP and changes GP subcellular localization, the type of ubiquitin linkage that signals GP localization to the nucleus is unknown. Malin ubiquitinates several enzymes involved in glycogen metabolism, including glycogen synthase, protein targeting to glycogen, glycogen debranching enzyme, and laforin to modulate glycogen architecture (7). It is interesting to note that ubiquitin addition by malin does not always result in proteasomal degradation, as is the case with GP. We hypothesize that the types of ubiquitin linkage differentiate the fate of the ubiquitinated glycogen metabolic enzymes. Additionally, whether the activity and re-export of GP rely on the removal of ubiquitin remains an open question. We speculate that similar to malin, a GP-specific deubiquitinase exists that can remove the ubiquitin so that GP is transported back to the cytoplasm (Figure 1A). Our lab is currently screening for the deubiquitinase associated with GP as well as identifying the type of ubiquitin linkage on GP. These findings will also elucidate other critical questions about the dynamics of nuclear trafficking and GP activity.
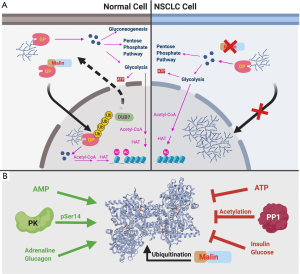
GP is a highly studied enzyme whose activity is modulated by multiple allosteric regulators—i.e., glucagon, insulin, ATP and AMP—and post-translational modifications—i.e., phosphorylation, acetylation, and now ubiquitination (Figure 1B). It will be important to determine how ubiquitination is integrated with the other post-translational modifications that impact GP activity. Equally important is defining how GP ubiquitination impacts the localization and activity of other glycogen regulating enzymes. It is possible that those enzymes also undergo shuttling in and out of the nucleus via ubiquitin modification. Defining these orchestrated events will allow us to elucidate the mechanisms by which nuclear glycogen is regulated.
Ferron also postulates concerning the multiple sources of acetyl-CoA in the cell and their fate in the metabolome. Ferron suggests that manipulating substrate availability for histone acetylation could have adverse consequences for the metabolome as a whole. Acetate and pyruvate are primary precursor metabolites for acetyl-CoA production, which are required for histone acetylation (8). Both metabolites exist in multiple cellular compartments, such as the nucleus, cytosol, and the mitochondria (9). In addition to nuclear glycogenolysis, pyruvate and acetate can be produced from glycolysis of glucose, oxidation of fatty acids, branched chain amino acid catabolism, and citric acid metabolism (9). Enzymes responsible for these reactions also exist in multiple compartments (10). For example, ATP citrate lyase, which converts acetyl-CoA from citrate, exists in both the cytosol and the nucleus (11). To this end, the nucleus and cytosol maintain separate pools of acetate for compartmentalized regulation (12). These results demonstrate the metabolic plasticity of cells to adapt to alternative biochemical pathways without altering the entire metabolome. While there are transporters that regulate substrate levels in the nucleus, it is difficult to determine the percent contribution of each substrate to histone acetylation. We anticipate that complex regulatory mechanisms govern the metabolic adaptation to histone acetylation. We propose that flux modeling and multi-Omic integration coupled to machine learning will shed light on these regulatory mechanisms.
Understanding the activity and role of malin in regulating glycogen metabolism will provide critical insights into the signaling crosstalk between cytosolic and nuclear metabolism. Our study suggests that down regulation of malin is a key signaling event that affects tumorigenesis. Prior studies have shown that the promoter region of malin undergoes extensive methylation in both NSCLC and in the aging populations (13,14). Therefore, it is likely that suppressed malin level is critical in the initial development of NSCLCs. As malin levels decrease, this would impact the accessibility of nuclear glycogen, further reducing the availability of carbon source for histone acetylation and creating a negative feedback loop. These findings place malin as a pivotal enzyme in tumorigenesis and potentially in aging.
Acknowledgments
We thank all members of the Gentry and Sun labs for valuable feedback.
Footnote
Conflicts of Interest: MS Gentry and RC Sun report personal fees and non-financial support from Maze Therapeutics and Valerion Therapeutics that had no role in this study. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sun RC, Dukhande VV, Zhou Z, et al. Nuclear Glycogenolysis Modulates Histone Acetylation in Human Non-Small Cell Lung Cancers. Cell Metabolism 2019;30:903-16.e7. [Crossref] [PubMed]
- Young LEA, Sun R, Emanuelle S, et al. Compartmentalized glycogenolysis regulates lung cancer transcription. FASEB J 2019;33:abstr 652.25.
- Zois CE, Harris AL. Glycogen metabolism has a key role in the cancer microenvironment and provides new targets for cancer therapy. J Mol Med (Berl) 2016;94:137-54. [Crossref] [PubMed]
- Terashima M, Fujita Y, Togashi Y, et al. KIAA1199 interacts with glycogen phosphorylase kinase β-subunit (PHKB) to promote glycogen breakdown and cancer cell survival. Oncotarget 2014;5:7040. [Crossref] [PubMed]
- Schnier JB, Nishi K, Monks A, Gorin FA, Bradbury EM. Inhibition of glycogen phosphorylase (GP) by CP-91,149 induces growth inhibition correlating with brain GP expression. Biochem Biophys Res Commun 2003;309:126-34. [Crossref] [PubMed]
- Lee WNP, Guo P, Lim S, et al. Metabolic sensitivity of pancreatic tumour cell apoptosis to glycogen phosphorylase inhibitor treatment. Br J Cancer 2004;91:2094-100. [Crossref] [PubMed]
- Gentry MS, Guinovart JJ, Minassian BA, et al. Lafora disease offers a unique window into neuronal glycogen metabolism. J Biol Chem 2018;293:7117-25. [Crossref] [PubMed]
- Liu X, Cooper DE, Cluntun AA, et al. Acetate is generated de novo from glucose metabolism in mammals and is coupled to central carbon metabolism. bioRxiv 2018.
- Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 2020;11:102. [Crossref] [PubMed]
- Huangyang P, Li F, Lee P, et al. Fructose-1, 6-Bisphosphatase 2 Inhibits Sarcoma Progression by Restraining Mitochondrial Biogenesis. Cell metabolism 2019. [PubMed]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009;324:1076-80. [Crossref] [PubMed]
- Bulusu V, Tumanov S, Michalopoulou E, et al. Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep 2017;18:647-58. [Crossref] [PubMed]
- Li Y, Gu J, Xu F, Zhu Q, Ge D, Lu C. Novel methylation-driven genes identified as prognostic indicators for lung squamous cell carcinoma. Am J Transl Res 2019;11:1997-2012. [PubMed]
- Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell 2015;14:924-32. [Crossref] [PubMed]


