
Forkhead box K1 regulates the malignant behavior of gastric cancer by inhibiting autophagy
Introduction
Gastric cancer (GC) is among the most prevalent forms of cancer and remains the second leading cause of cancer-related death (1). Recently, treatments for GC have substantially improved; however, due to a variety of genetic mutations and abnormal signaling pathways underlying GC progression, GC mortality remains high (2). A better understanding of the unique molecular patterns of GC development will help researchers identify the mechanisms underlying alterations in tumor activities.
Forkhead box (FOX) transcription factors are involved in tumor development (3), embryogenesis (4) and the regulation of various physiological processes, such as cell survival (5), oxidative stress (6), and control of lifespan (7). Forkhead box K1 (FOXK1) is a member of the FOX family of transcription factors and is commonly studied in myogenic cells (8). Recently, abnormal expression of FOXK1 was suggested to contribute to tumor development. According to Li et al. (9), FOXK1 regulates p21 expression and promotes the proliferation and metastasis of ovarian cancer. Haitao Xu and other scholars postulated that FOXK1 promotes glioblastoma proliferation and metastasis by inducing snail transcription (10). However, the link between FOXK1 and autophagy in GC remains unclear.
Autophagy is a process that is highly conserved throughout many organisms, but there is controversy regarding whether autophagy is associated with cell death. Autophagy inhibits tumor growth in the early stages by removing damaged organelles or proteins; however, in advanced tumors, autophagy elicits the opposite effect (11). In GC, downregulation of long noncoding RNA LINC01419 inhibits tumor cell migration and invasion and tumor growth. This phenomenon is promoted by inactivation of the PI3K/AKT1/mTOR pathway (12). At the same time, studies have shown that FOXK1 can regulate the AKT/mTOR pathway in liver cell lines (13), which suggests that autophagy and the PI3K/AKT/mTOR pathway play a key role in the malignant behavior of GC, but the role of FOXK1 in this dynamic is still unknown.
In this study, increased FOXK1 expression was significantly correlated with progression, metastasis, and adverse outcomes in patients with GC. In addition, this study revealed for the first time that FOXK1 promotes the malignant behavior of GC by inhibiting autophagy.
Methods
Cells and culture conditions
Four human GC cell lines (SGC7901, BGC823, MGC803, and HGC27) as well as the immortalized gastric mucosal cell line GES1 were obtained from the Cell Bank of the Chinese Academy of Sciences. AGS GC cells were obtained from Zhongqiao New Prefecture in Shanghai. All cells were grown in medium containing 10% fetal bovine serum (FBS; Gibco, NY, USA) and 1% penicillin-streptomycin (HyClone, Logan, UT, USA) in a standard humidified incubator.
For experiments involving 3-MA (MedChemExpress, Shanghai, China), cells were pretreated with 200 µM 3-MA for 4 h before transfection.
Gene expression analysis
We utilized Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/detail.php?gene=FOXK1) to compare gene expression in tumor and healthy tissues. We used Kaplan-Meier Plot (http://kmplot.com/analysis/index.php?p=service&cancer=gastric) to analyze the correlation between overall survival (OS) and FOXK1 expression in patients with GC and Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/), a public biological resource, for pathway analysis.
Patients and specimens
We assessed 43 pairs of freshly frozen primary GC and matched healthy tissue samples collected at the Affiliated Hospital of Qingdao University. Samples from patients with GC from September 2016 to October 2018 who did or did not receive chemotherapy were collected for pathological assessment and analysis of progression. All individual patients were informed about the goals of this study and provided written informed consent.
Tissue microarrays (TMAs)
GC tissues and matched noncancerous stomach tissues were analyzed using TMAs purchased from AMOS Scientific (Beijing, China). Forty-three tissue pairs in the TMAs were subjected to immunohistochemistry to assess FOXK1 expression.
The TMAs were independently evaluated by two investigators. The staining intensity was scored from 0 to 3 points as follows: 0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining. The percentage of positive cells was scored as follows: 0, <10% positive cells; 1, 10–35% positive cells; 2, 36–70% positive cells; and 4, >70% positive cells.
Cell transfection
The FOXK1 open reading frame and 3'-untranslated region were previously cloned into pcDNA 3.1(+). AGS and MGC803 cells were cultured to 80–90% confluence and transfected with the pcDNA3.1-FOXK1 plasmid (Genechem Co., Ltd., Shanghai, China) or FOXK1 siRNA (Ribobio Co., Guangzhou, China) (Table S1) using Lipofectamine 3000 (Life Technologies, Shanghai, China) according to the manufacturer’s protocols. At 48 h after transfection, the experiments were conducted.
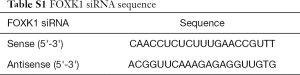
Full table
RNA isolation and quantitative real-time RT-PCR
Total RNA was extracted using TRIzol Reagent (TaKaRa, Beijing, China) and reverse transcribed into cDNA using the PrimeScript RTMaster Mix (Perfect Real Time) reagent (TaKaRa, Beijing, China) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed on an ABI 7500HT Fast Real-Time PCR System (Applied Biosystems, CA, USA). The 10 µL reaction mixture consisted of SYBR GREEN PCR Master Mix (TaKaRa, Beijing, China), 500 nmol of primers and 300 ng of cDNA templates. The reaction was initially denatured at 95 °C for 5 min and then subjected to 60 cycles of 94 °C for 20 s, 60 °C for 20 s and 72 °C for 40 s. Next, the reaction temperature was increased from 72 °C to 95 °C, and the reactions were finally extended for 5 min at 72 °C before the temperature was increased at a rate of 0.1 °C/s for continuous fluorescence acquisition. Each qRT-PCR assay was repeated three times, and the cDNAs were measured in duplicate. The average fold of relative mRNA expression was determined using the 2−ΔΔCt method with GAPDH as an internal control. Primer sequences for qRT-PCR were as follows: FOXK1, forward 5'-GCCACAAAGGCTGGCAGAATT-3' and reverse 5'-TGGCTTCAGAGGCAGGGTCTAT-3'; class I PI3K, forward 5'-AACGAGAACGTGTGCCATTTG-3' and reverse 5'-AGAGATTGGCATGCTGTCGAA-3'; class III PI3K, forward 5'-AGAGTTATGCGTTCTTT-GCTGGC-3' and reverse 5'-GGGGTTCTAAAGGCAA-CGGGATA-3'; and GAPDH, 5'-TCGACAGTCA-GCCGCATCTT-3' and reverse: 5'-GAGTTAAAAGCAG-CCCTGGTG-3'.
Western blot assays
Total protein was extracted from GC cells and tumor tissues collected from nude mice. Proteins were subsequently separated on SDS-PAGE gels and then transferred to nitrocellulose membranes, which were blocked with 5% skim milk for 1 h at room temperature (RT). Primary antibodies (listed in Table S2) were incubated with the membranes overnight at 4 °C. The membranes were then washed three times with phosphate-buffered saline containing Tween 20 (PBST) for 15 min per wash and incubated with HRP-conjugated secondary antibodies at RT for 2 h. After additional washes, the electrochemiluminescence (ECL) (Life Technologies, Shanghai, China) of the bands was detected using a Bio-Rad infrared gel imaging system (ChemiDoc XRS+).

Full table
5-ethynyl-2’-deoxyuridine (EdU) incorporation assay
Tumor cells were seeded on 24-well plates (1×105 cells/well), cultured for 48 h, and incubated with medium containing 50 µM EdU (Beyotime, Shanghai, China) for 2 h. Cells were then fixed with 4% paraformaldehyde (Beyotime, Shanghai, China) and permeabilized, after which a click reaction mixture was added to the cells (200 µL/well) for 30 min. Hoechst 33342 (200 µL/well) staining for 30 minutes was performed to stain nuclei, and cells were visualized under a fluorescence microscope.
Transwell migration and wound healing assays
Cell migration and invasion were detected using Transwell chambers (BD, Franklin Lakes, NJ, USA) without and with a Matrigel coating, respectively, according to the manufacturer’s instructions. After transfection, 2×105 cells in serum-free medium were added to the upper chamber of the inserts, and medium containing 20% FBS was added to the lower chamber. After 24 h of culture, cells were fixed with 4% paraformaldehyde for 30 min, stained with crystal violet for 20 min and washed with phosphate-buffered saline (PBS). The cells were then counted under a microscope. The average number of cells in five fields of view on each membrane was counted three times.
For the wound healing assay, cells were plated in a 6-well plate, and then a scratch was created through a confluent cell layer with a sterile pipette tip. Debris was removed by washing the wells with PBS, and the cells were imaged immediately after scratching (baseline) and every 24 h thereafter.
Laser confocal microscopy
AGS and MGC803 cells treated with different compounds were seeded on coverslips in 24-well plates. Cells were fixed with 4% paraformaldehyde, incubated with 5% bovine serum albumin (BSA) (Sigma, Shanghai, China) for 30 min and stained with DAPI (Beyotime, Shanghai, China) for 7 min. The slides were air dried and sealed with a sealing tablet. LC3 fluorescence was observed under a confocal laser microscope, and the number of LC3 spots was counted in 3 independent replicates.
Transmission electron microscopy
Treated cells were trypsinized and collected after centrifugation. Then, they were fixed overnight with 2.5% phosphate-buffered glutaraldehyde (Sinopharm Chemical Reagents Co., Shanghai, China), after which they were embedded, sectioned, double stained with uranyl acetate and lead citrate and analyzed under a transmission electron microscope.
Establishment of tumor xenografts in CB17/SCID mice
In all, 3×106 cells in 100 µL of PBS were subcutaneously injected into the right flanks of mice. The tumor growth rates were monitored by measuring the tumor diameter every 7 days, and the tumor volume was calculated using the following equation: 1/2 LW2, where L=length and W=width. The mice were treated with the indicated compounds 28 days later, and tumors were collected after gravimetric analysis.
Statistical analysis
SPSS 19.0 software (SPSS Inc., IL, USA) was employed for all statistical analyses. Data are presented as the means ± S.D. and were compared using Student’s t-test or analysis of variance. P<0.05 was considered significant.
Results
High FOXK1 expression predicts a poor prognosis for patients with GC
FOXK1 expression in healthy stomach tissues and GC tissues was compared using GEPIA (Figure 1A), and we observed a significant increase in FOXK1 expression in GC tissues compared to that in healthy tissues. Additionally, the OS (Figure 1B) data obtained from the Kaplan-Meier plot were consistent with our curve created from data obtained from The Cancer Genome Atlas (TCGA), indicating that FOXK1 expression is associated with the survival of patients with GC. We next assessed FOXK1 expression in a TMAs comprising 43 pairs of healthy control and GC tumor tissues from patients (Figure 1C). The tissue specimens were divided into a low FOXK1 expression group [immunoreactive score (IRS): 0–4] and a high FOXK1 expression group (IRS: 4–12) based on a quantitative analysis of protein staining. We identified a correlation between the high FOXK1 expression group and the malignant progression of GC tissues (Figure 1D,E, P<0.05), as high FOXK1 expression was observed in 37 of 43 (86.0%) GC tissues (Table 1).
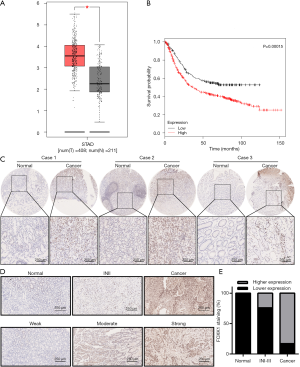
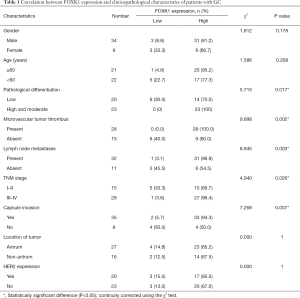
Full table
The clinical correlations between FOXK1 expression and the clinicopathological parameters of GC were further analyzed to explore the significance of FOXK1 upregulation (Table 1). High levels of FOXK1 were correlated with specific GC clinicopathological characteristics: pathological differentiation (P=0.017), microvascular tumor thrombus (P=0.002), lymph node metastases (P=0.003), tumor, node, metastasis (TNM) stage (P=0.026), and capsule invasion (P=0.007). FOXK1 expression was not correlated with sex, age, tumor location, or HER2 expression in the tumor. Taken together, these data clearly confirmed FOXK1 upregulation in GC tissues and the correlation of this increased expression with the degree of malignancy and clinical prognosis.
The effects of FOXK1 on GC cell proliferation
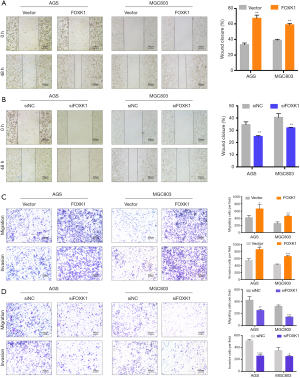
Since elevated FOXK1 expression is correlated with malignant progression, FOXK1 may play an important role in one or more steps of GC proliferation. First, we further verified the increase in FOXK1 expression in GC cell lines using western blotting (Figure 2A). Increased protein levels of FOXK1 were detected in the AGS, SGC7901, BGC823, MGC803, and HGC27 cell lines compared to the normal human gastric cell line GES-1 (used as a control). Based on these results, we selected the AGS and MGC803 cell lines for further study because they exhibited a moderate increase in FOXK1 levels. AGS and MGC803 cells transfected with the pcDNA3.1-FOXK1 plasmid (FOXK1) were subsequently used in a proliferation assay, with cells transfected with an empty vector (vector) serving as a control. We also knocked down FOXK1 expression in AGS and MGC803 using an siRNA (Table S1) and compared its effects to those of control siRNAs. The transfection efficiency was validated using real-time PCR and western blotting (Figure 2B,C). Next, the results of the EdU assays suggested that FOXK1 overexpression markedly accelerated the growth of AGS and MGC803 cells, whereas downregulation of FOXK1 expression significantly reduced AGS and MGC803 proliferation (Figure 2D). Based on these findings, we can surmise that FOXK1 increases GC cell proliferation.
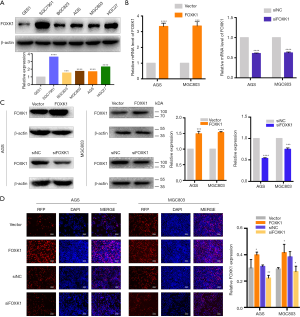
FOXK1 affects GC cell migration and invasion
Because previous studies have reported a close association between FOXK1 expression and clinicopathological features, we evaluated the effects of FOXK1 on GC cell motility in vitro. Wound healing and Transwell assays were utilized to determine how FOXK1 influences GC cell migration and invasion. Based on the results from the wound healing assays, FOXK1 overexpression led to increased motility compared with that of the control cells (Figure 3A). In contrast, FOXK1 knockdown in these same cells led to slower wound closure (Figure 3B). In Transwell assays, FOXK1 overexpression increased AGS and MGC803 cell migration and invasion compared with those of cells transfected with vector controls (Figure 3C). FOXK1 knockdown effectively reduced the number of invading cells in both cell lines (Figure 3D). FOXK1 thus drives the migration and invasion of GC cells in vitro.
FOXK1 suppresses autophagy in GC
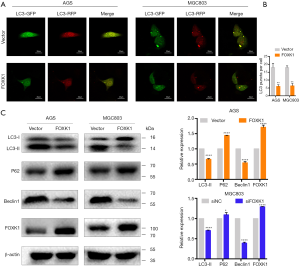
Autophagy is closely linked to oncogenesis (14). Strategies that induce autophagy have been reported to inhibit tumor proliferation, invasion and migration (15,16). However, the relationship between FOXK1 expression and autophagy has not been previously reported in GC. To elucidate the relationship between FOXK1 and autophagy, autophagy activity was assessed using a confocal laser imaging system. An emerging approach was used to analyze autophagosome-mediated dynamic changes in the protein levels and protein degradation. The functional switching of yellow fluorescence to red fluorescence reflects the functional autophagy flux after LC3 is fused in tandem with acid-resistant mCherry (such as RFP) and acid-sensitive GFP. As shown in Figure 4A,B, the yellow fluorescence of autophagic puncta was dramatically reduced in cells transfected with the pcDNA3.1-FOXK1 plasmid. Western blotting confirmed clear reductions in the expression levels of the autophagy-associated proteins LC3-II and Beclin1, as well as increased levels of p62 (Figure 4C).
FOXK1 knockdown promotes autophagy in GC cells
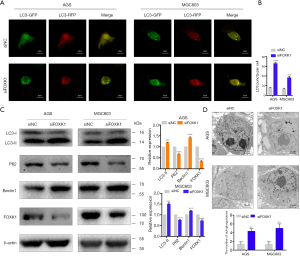
We assessed autophagy in cells with FOXK1 knockdown to further confirm the effect of FOXK1 on autophagy. The results from the confocal laser analysis revealed a significant increase in autophagy following FOXK1 knockdown (Figure 5A,B), and western blotting confirmed clear increases in LC3-II and Beclin1 levels and decreased p62 levels (Figure 5C). Furthermore, transmission electron microscopy revealed autophagic vesicles containing engulfed organelles in the cytoplasm (Figure 5D).
FOXK1 induces autophagy in GC cells by modulating the PI3K/AKT/mTOR signaling pathway
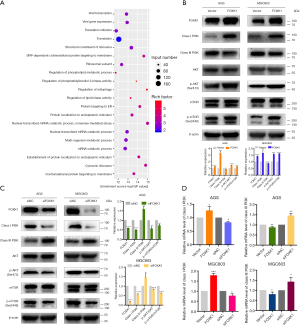
The PI3K/AKT pathway was recently shown to function as a negative regulator of autophagy (17). Therefore, we used KEGG signaling pathway analysis to identify pathways that FOXK1 regulates and found that FOXK1 regulates PI3K activation (Figure 6A). Several other studies have shown that class I PI3K and class III PI3K have opposing functions in autophagy regulation (18). Therefore, we observed the effects of overexpression and knockdown of FOXK1 expression on class I PI3K, class III PI3K, AKT, and mTOR in GC cells. Western blots showed that levels of class I PI3K, the p-AKT/total AKT ratio and the p-mTOR/total mTOR ratio were significantly increased and that expression of class III PI3K was decreased after FOXK1 overexpression (Figure 6B). In addition, FOXK1 knockdown led to the opposite trend (Figure 6C). We also used real-time PCR to verify that FOXK1 can increase mRNA expression in class I PI3K, whereas the expression of class III PI3K is reduced (Figure 6D). Taken together, these data indicate that FOXK1 regulates autophagy at least in part through the PI3K/AKT/mTOR pathway. At the same time, FOXK1 inhibits autophagy by upregulating class I PI3K and downregulating class III PI3K.
FOXK1 knockdown increases autophagy inhibited by 3-methyladenine (3-MA)
3-MA, a class III phosphatidylinositol 3-kinase (PI3K) inhibitor, is currently the most widely used autophagy inhibitor (19). We transfected AGS and MGC803 cells with siNC and siFOXK1 to further determine the role of FOXK1 in the proliferation and invasion of cells in which 3-MA (200 µm) suppressed autophagy. As shown in Figure 7A, in the presence or absence of 3-MA, LC3-II expression was increased and p62 expression was decreased after treatment with siFOXK1. Furthermore, western blotting results confirmed that the downregulation of LC3-II caused by 3-MA was enhanced by siFOXK1 in AGS and MGC803 cells. In contrast, upregulation of p62 induced by 3-MA was alleviated by siFOXK1. In addition, the results of the EdU and Transwell assays showed that silencing FOXK1 attenuated 3-MA-induced proliferation and invasion (Figure 7B,C). Based on these results, FOXK1 plays an important role in 3-MA-mediated suppression of autophagy in GC cells.
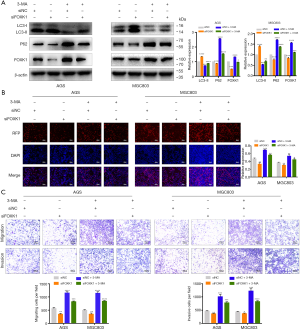
Downregulation of FOXK1 inhibits GC growth in vivo
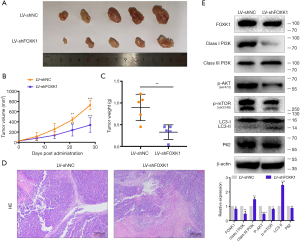
We established mouse xenograft models to further examine the role of FOXK1 in vivo. First, LV-shNC and LV-shFOXK1 were transfected into MGC803 GC cells. Next, equal numbers of both groups of MGC803 cells were injected into the hind limbs of 4-week-old CB17/SCID mice. Four weeks after implantation, all the mice were sacrificed, and the tumor size and volume were recorded. As shown in Figure 8A, the tumors were significantly smaller in mice inoculated with LV-shFOXK1-transfected GC cells than in those inoculated with LV-shNC-transfected cells. Fifteen days after inoculation, the growth curve of the control group was significantly more pronounced than that of the LV-shFOXK1 group (Figure 8B). Accordingly, the xenografted tumors from the mice were weighed, and the tumors from the LV-shNC group were significantly heavier than those from the LV-shFOXK1 group (Figure 8C). As shown in Figure 8D, hematoxylin and eosin (H&E) staining confirmed the apparent heterogeneity of the cells in the tumor, with large nuclei and single staining. At the same time, western blotting analysis showed significantly reduced levels of FOXK1, class I PI3K, p-AKT, p-mTOR and p62 in tumors from mice in the LV-shFOXK1 group, which was accompanied by an increase in the level of class III PI3K and LC3-II (Figure 8E). Taken together, these data suggest that inhibiting FOXK1 expression increases autophagy levels in GC through the PI3K/AKT/mTOR pathway.
Discussion
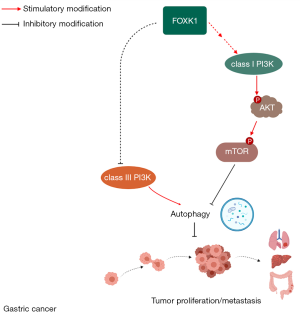
In this study, we analyzed the relationship between FOXK1 expression and clinicopathological features of GC patients. FOXK1 expression is closely correlated with the degree of GC malignancy. Subsequently, we determined that FOXK1 significantly affects the proliferation, invasion, and metastasis of GC cells and that autophagy may play a role in this process. Finally, FOXK1 was shown to activate the class I PI3K/AKT/mTOR pathway and inhibit class III PI3K, both of which block autophagy. Overall, FOXK1 represents a potential prognostic marker and a positive predictor of GC growth and invasiveness. Its upregulation promotes the malignant behavior of GC by inhibiting autophagy, and its mechanism may be related to the regulation of the PI3K/AKT/mTOR signaling pathway (Figure S1).
The FOX family of transcription factors is involved in tumorigenesis, tumor progression and embryogenesis (20,21). Based on accumulating evidence, FOXK1, a member of the FOX family, functions as an oncogene in many cancers when it is overexpressed. Wu et al. (22) also confirmed that FOXK1 interacts with FHL2 to promote colon cancer proliferation, invasion and metastasis. The abnormal expression of FOXK1 and consequent activation of downstream signaling pathways have been shown to lead to tumorigenesis and progression and alter the tumor phenotype. However, the mechanism by which FOXK1 promotes the malignant behavior of GC remains unclear. Our results confirm for the first time that FOXK1 overexpression increases proliferation and drives invasion and metastasis by inhibiting autophagy. Therefore, FOXK1 overexpression may be an important molecular target for cancer growth and metastasis, in which autophagy plays a key role.
Autophagy is a dynamic physiological process that aims to maintain cellular homeostasis. In some contexts, this process promotes cell survival by generating energy; however, this process changes drastically in tumors (23). Cancer cell-induced autophagy is more likely to be a dynamic mechanism that supports or inhibits tumor cell survival. Chen HT and other scholars concluded that autophagy can act as a cancer suppressor and tends to prevent metastasis by selectively downregulating key transcription factors of EMT (24). According to a study by Kroemer, stress-induced hyperautophagy inhibits cell survival (16). As shown in a study by Wei R, inhibiting autophagy promotes the invasion and metastasis of colon cancer cells (15). Thus, autophagy represents a physiological process that can inhibit the proliferation and migration of tumor cells. Based on our results, FOXK1 overexpression decreases the LC3-II/LC3-I ratio and Beclin1 levels and increases p62 levels. After FOXK1 was knocked down, the LC3-II/LC3-I ratio and Beclin1 levels increased and p62 levels decreased, indicating that FOXK1 is a key factor regulating autophagy. This result is consistent with the findings reported by Christopher John Bowman (25). In the present study, the proliferation, invasion and metastasis of GC cell lines were increased after 3-MA-mediated inhibition of autophagy. Thus, FOXK1 may regulate autophagy during the later stages of tumor development, thereby promoting the malignant behaviors of GC.
The PI3K/AKT/mTOR signaling pathway plays a pivotal role in both development and disease, with a known role in cancer (26,27). PI3K is a member of a family of conserved lipid kinases whose aberrant activation is frequently observed in human cancers (28). Class I PI3K, which negatively regulate autophagy, phosphorylate PIP2 to produce PIP3. PIP3 recruits the second messenger AKT to send signals to mTOR to consequently inhibit autophagy (29,30). By contrast, class III PI3K drive processes that promote autophagy (18). In the present study, our results support the hypothesis that FOXK1 inhibits autophagy at least in part through the PI3K/AKT/mTOR signaling pathway to promote malignant behavior of tumors. Based on these findings, FOXK1 plays an important role in mediating GC progression, which provides new insights into further understanding and developing GC-based therapies.
Conclusions
In summary, FOXK1 inhibits autophagy by activating the PI3K/AKT/mTOR pathway and inhibiting class III PI3K activity, thereby promoting the malignant behaviors of GC. These data reveal a new molecular mechanism of FOXK1 and autophagy in GC. In addition, FOXK1 may be a key factor predicting tumor progression and clinical outcomes in patients with GC. Therefore, the FOXK1-autophagy axis represents a potential novel target for treating GC.
Acknowledgments
We would like to thank M. Z. and Y. Q. for providing constructive suggestions and participating in discussions.
Funding: This study was supported by the Key Research and Development Foundation Projects of Shandong Province (2017GSF218109) and the Science and Technology Foundation Projects of Shandong Province (J15LL58).
Footnote
Conflicts of Interest: The authors declare no conflicts of interest.
Ethical Statement: The authors are accountable for all aspects of this work in ensuring that questions related to the accuracy or integrity of any part of this work are appropriately investigated and resolved. The Medical Ethics Committee of Qingdao University and the Affiliated Hospital of Qingdao University approved the collection of clinical materials for research purposes. All samples were collected and analyzed after obtaining written informed consent from each patient. All animal experiments adhered to the Principles of Care and Use of Laboratory Animals of Qingdao University and were approved by the Subcommittee on the Ethics of Experimental Animal Welfare within the Medical Ethics Committee of Qingdao University (approval no. AHQDMAL20181201, approval date: 1 December 2018).
References
- Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J 2014;55:621-8. [Crossref] [PubMed]
- Lazăr DC, Tăban S, Cornianu M, et al. New advances in targeted gastric cancer treatment. World J Gastroenterol 2016;22:6776-99. [Crossref] [PubMed]
- Wang J, Li W, Zhao Y, et al. Members of FOX family could be drug targets of cancers. Pharmacol Ther 2018;181:183-96. [Crossref] [PubMed]
- Wolf I, Bose S, Williamson EA, et al. FOXA1: Growth inhibitor and a favorable prognostic factor in human breast cancer. Int J Cancer 2007;120:1013-22. [Crossref] [PubMed]
- Sturgill TW, Stoddard PB, Cohn SM, et al. The promoter for intestinal cell kinase is head-to-head with F-Box 9 and contains functional sites for TCF7L2 and FOXA factors. Mol Cancer 2010;9:104. [Crossref] [PubMed]
- Ma J, Matkar S, He X, et al. FOXO family in regulating cancer and metabolism. Semin Cancer Biol 2018;50:32-41. [Crossref] [PubMed]
- Fu W, Ma Q, Chen L, et al. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J Biol Chem 2009;284:13987-4000. [Crossref] [PubMed]
- Shi X, Wallis AM, Gerard RD, et al. Foxk1 promotes cell proliferation and represses myogenic differentiation by regulating Foxo4 and Mef2. J Cell Sci 2012;125:5329-37. [Crossref] [PubMed]
- Li L, Gong M, Zhao Y, et al. FOXK1 facilitates cell proliferation through regulating the expression of p21, and promotes metastasis in ovarian cancer. Oncotarget 2017;8:70441-51. [PubMed]
- Xu H, Huang S, Zhu X, et al. FOXK1 promotes glioblastoma proliferation and metastasis through activation of Snail transcription. Exp Ther Med 2018;15:3108-16. [PubMed]
- Qian HR, Yang Y. Functional role of autophagy in gastric cancer. Oncotarget 2016;7:17641-51. [Crossref] [PubMed]
- Wang LL, Zhang L, Cui XF. Downregulation of long noncoding RNA LINC01419 inhibits cell migration, invasion, and tumor growth and promotes autophagy inactivation of the PI3K/Akt1/mTOR pathway in gastric cancer. Ther Adv Med Oncol 2019;11:1758835919874651. [Crossref] [PubMed]
- Cui H, Gao Q, Zhang L, et al. Knockdown of FOXK1 suppresses liver cancer cell viability by inhibiting glycolysis. Life Sci 2018;213:66-73. [Crossref] [PubMed]
- Galluzzi L, Pietrocola F, Bravo-San Pedro JM, et al. Autophagy in malignant transformation and cancer progression. EMBO J 2015;34:856-80. [Crossref] [PubMed]
- Wei R, Xiao Y, Song Y, et al. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J Exp Clin Cancer Res 2019;38:112. [Crossref] [PubMed]
- Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol cell 2010;40:280-93. [Crossref] [PubMed]
- Maejima Y, Kyoi S, Zhai P, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med 2013;19:1478-88. [Crossref] [PubMed]
- Li J, Zhou J, Zhang D, et al. Bone marrow-derived mesenchymal stem cells enhance autophagy via PI3K/AKT signalling to reduce the severity of ischaemia/reperfusion-induced lung injury. J Cell Mol Med 2015;19:2341-51. [Crossref] [PubMed]
- You P, Wu H, Deng M, et al. Brevilin A induces apoptosis and autophagy of colon adenocarcinoma cell CT26 via mitochondrial pathway and PI3K/AKT/mTOR inactivation. Biomed Pharmacother 2018;98:619-25. [Crossref] [PubMed]
- Pellegrini P, Strambi A, Zipoli C, et al. Acidic extracellular pH neutralizes the autophagy-inhibiting activity of chloroquine: implications for cancer therapies. Autophagy 2014;10:562-71. [Crossref] [PubMed]
- Teh MT, Wong ST, Neill GW, et al. FOXM1 Is a Downstream Target of Gli1 in Basal Cell Carcinomas. Cancer Res 2002;62:4773. [PubMed]
- Wu M, Wang J, Tang W, et al. FOXK1 interaction with FHL2 promotes proliferation, invasion and metastasis in colorectal cancer. Oncogenesis 2016;5:e271. [Crossref] [PubMed]
- Levy JM, Towers CG, Thorburn A. Targeting Autophagy in Cancer. Nature reviews Cancer 2017;17:528-42. [Crossref] [PubMed]
- Chen HT, Liu H, Mao MJ, et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol Cancer 2019;18:101. [Crossref] [PubMed]
- Bowman CJ, Ayer D, Dynlacht B. Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Nat Cell Biol 2014;16:1202-14. [Crossref] [PubMed]
- Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet 1999;21:99-102. [Crossref] [PubMed]
- Levine DA, Bogomolniy F, Yee CJ, et al. Frequent Mutation of the PIK3CA Gene in Ovarian and Breast Cancers. Clin Cancer Res 2005;11:2875-8. [Crossref] [PubMed]
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene 2008;27:5497-510. [Crossref] [PubMed]
- Saleh SN, Albert AP, Large WA. Activation of native TRPC1/C5/C6 channels by endothelin-1 is mediated by both PIP3 and PIP2 in rabbit coronary artery myocytes. J Physiol 2009;587:5361-75. [Crossref] [PubMed]
- van Rheenen J, Song X, van Roosmalen W, et al. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol 2007;179:1247-59. [Crossref] [PubMed]


