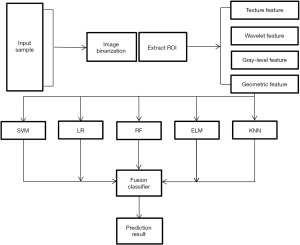
Multiclassifier fusion based on radiomics features for the prediction of benign and malignant primary pulmonary solid nodules
Introduction
Pulmonary nodules usually refer to round or irregular lesions in the lung that are no more than 3 cm in diameter. With the advancement of spiral computed tomography (CT) scanning, reconstruction technology, and low-dose chest CT screening, the detection rate of pulmonary nodules is increasing. However, since the same kind of shadow can be cast by different diseases and different shadows can be cast by the same disease, benign and malignant nodules can be confused. In 2012, Dutch scholar Lambin et al. (1) first proposed the concept of radiomics. Kumar et al. (2) defined it as “high-throughput extraction and analysis of a large number of advanced quantitative imaging features from CT, magnetic resonance imaging (MRI), and positron emission tomography (PET)”. In recent years, radiomics has played an important role in the identification of benign and malignant lesions, judgment of the malignancy of tumors, selection of treatment methods, and monitoring of therapeutic effect, and it has guiding significance for the development of personalized treatment plans (3-6). In this study, based on traditional imaging differential diagnosis, a prediction model of benign and malignant pulmonary solid nodules based on radiomics was constructed using a feature selection algorithm combined with multiple classifiers, and the prediction performance of the classifiers was quantitatively evaluated.
Methods
Clinical information
The clinical, pathological and imaging information of 342 patients with pulmonary solid nodules confirmed by histopathology or follow-up in the Sir Run Run Shaw Hospital affiliated with the School of Medicine of Zhejiang University and Yinzhou Hospital affiliated with the School of Medicine of Ningbo University from January 2015 to December 2018 were retrospectively analyzed. The inclusion criteria were (I) isolated solid nodules in the lung were identified on chest CT examination, and each nodule had a clear and complete thin layer to reconstruct the data in Digital Imaging and Communications in Medicine (DICOM) format; (II) diagnosis was confirmed by histopathology or follow-up after clinical treatment; (III) the reconstructed thin-layer image had no obvious calcification and/or fat content; (IV) there was no history of extrapulmonary malignancies; (V) it was untreated before CT examination.
CT examination
The CT scan was performed using Siemens Definition AS 64 and Philips Brilliance 64-row multilayer spiral CT scanner. The scanning range was from the thorax entrance to the adrenal level. The scanning parameters were as follows: tube voltage of 120 kVp, tube current automatic adjustment technique, pitch of 1.2, collimation of 64×0.625 mm, reconstruction layer thickness/layer spacing of 1.25 mm/0.625 mm, matrix of 512×512, reconstruction convolution function of B70f, window width of 1,200 HU, window level of −600 HU, and all images exported in DICOM format.
Image analysis
Segmentation of region of interest (ROI) of nodules
Using ITK-SNAP software (Version 3.4.0, http://www.itksnap.org/), two radiologists with 5 years of experience manually and semiautomatically delineated the maximum boundary of the nodules layer by layer. Blood vessels and bronchus were avoided. The longest diameter of the nodule and the presence of the spicule sign, lobulation sign, vacuole sign, and vessel convergence sign were extracted and recorded (Figure 1A,B). The ROI segmentation was checked by one senior radiologist.
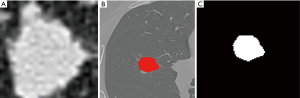
Image and data preprocessing
The images and data were preprocessed using image binarization and data normalization. Image binarization (Figure 1C) was done to set the gray value of each pixel on the images to 0 or 1. Binary images are conducive to further processing of the images so that image information other than the ROI can be eliminated to avoid the introduction of noise. In addition, in order to eliminate the influence of feature vectors of different dimensions on the analysis results, the extracted original radiomics feature data were standardized. All the feature vectors after processing were on the same order of magnitude and conformed to the standard normal distribution, that is, the mean was 0 and the standard deviation was 1. The conversion function was x* = (x−µ)/σ, where x, µ, and σ are the actual value, statistical average, and standard deviation of all feature vectors, respectively.
Radiomics feature extraction
The radiomics features of all segmented nodules were extracted using the Matlab2018b software (http://www.mathworks.com/) (MathWorks Co., USA). A total of 450 features in four major categories were extracted from all nodules, including geometric features, texture features, gray-level features, and wavelet features. Texture features included the common gray-level cooccurrence matrix (GLCM), gray-level run-length matrix (GLRLM), gray-level size zone matrix (GLSZM), neighborhood gray tone difference matrix (NGTDM), and neighborhood gray-level difference matrix (NGLDM). The local binary pattern (LBP) was also used to describe local texture features of the images.
Feature selection and model construction
A total of 342 samples were divided into training set, test set and verification set at a ratio of 7:2:1 using the random index method. The relief feature selection algorithm was used to screen the 450 radiomics features. After repetitive operations (an average of 10 repeats was used as the feature weight), 25 robust features with a major role in classification in the training set were screened. The classifiers were tested and verified based on the selected feature set, and a prediction model for distinguishing benign and malignant primary pulmonary solid nodules was constructed. The five classifiers were a support vector machine (SVM), random forest (RF), logistic regression (LR), extreme learning machine (ELM), and K-nearest neighbor (KNN). Finally, the weighted voting method (an algorithm that fuses classifiers (7) was used to fuse the prediction results of the above five classifiers, and each weight wi of the weighted fusion was calculated using the Lagrangian and QR decomposition method. The Lagrangian was used to construct the objective function, and QR decomposition was used to obtain the analytical solution. The classifier fusion method was as follows:
|
| [1] |
|
| [2] |
|
| [3] |
The Lagrangian expression was as follows:
|
| [4] |
|
| [5] |
where wi is the weight corresponding to the ith classifier,, are the QR decomposition of h(x), and .
Cross-validation of classifiers
The method of simple cross-validation (8) was used to verify the robustness of the classifiers. The process was as follows: (I) random integers 1 to 342 (the sample size was 342) were randomly generated. (II) The data set was divided into a training set, a test set, and a verification set at a ratio of 7:2:1 using the random number index method. (III) The five classifiers were trained, tested, and verified separately, and the training, testing, and verification results of the fusion classifier were obtained based on the results of the five classifiers. (IV) The process of (I) to (III) was repeated 10 times.
Statistics
SPSS 20.0 and Matlab2018b software were used for statistical analysis of the data in this study. Measurement data are expressed as mean ± standard deviation. Count data are expressed as a ratio or percentage. Differences in sex and imaging signs between the two groups were compared by the chi-squared test. The differences in age and the longest diameter of the lesions were compared using the two-independent-sample T test. P<0.05 was considered statistically significant. The diagnostic performance of the classifiers was described using precision, recall rate, and area under the receiver operating characteristic (ROC) curve (AUC).
Results
General information
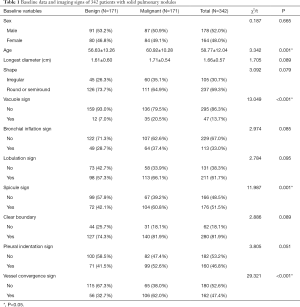
This study enrolled 342 patients with solid pulmonary nodules. Among them, 171 patients (91 males and 80 females) had benign nodules, with an average age of 56.63±13.26 years, and the longest diameter of nodules was 1.61±0.60 cm. Of them, 120 cases were confirmed by histology, including 34 cases of inflammatory pseudotumor (inflammatory granuloma), 31 cases of tuberculosis, 25 cases of fungus infection, 17 cases of hamartoma, and 13 cases of sclerosing hemangioma. The remaining 51 cases were confirmed by follow-up after treatment to have nodules that shrank or disappeared. There were 171 patients (87 males and 84 females) with malignant nodules, with an average age of 60.92±10.28 years and a longest nodule diameter of 1.71±0.54 cm, all of which were confirmed by histology, including 51 cases of squamous cell carcinoma, 75 cases of adenocarcinoma, 9 cases of large cell carcinoma, 24 cases of small cell carcinoma, 9 cases of adenosquamous carcinoma, and 3 cases of sarcomatoid carcinoma. The general information of the patients is shown in Table 1.
Full table
Weight distribution of radiomics features
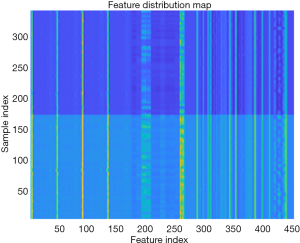
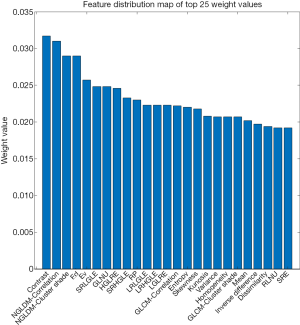
The entire experimental process is shown in Figure 2. The distribution of all 450 radiomics features between the two groups is shown in Figure 3. The features of benign vs. malignant nodules were significantly different, indicating that the extracted features objectively reflected the essential attributes of pulmonary nodules. The distribution of the top 25 features according to the weight after feature selection is shown in Table 2 and Figure 4. The features with the greatest weights were mainly concentrated in texture features, wavelet features, and gray-level features.
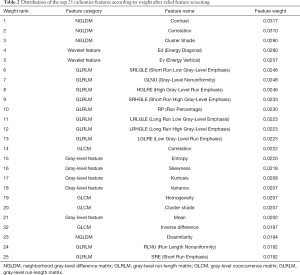
Full table
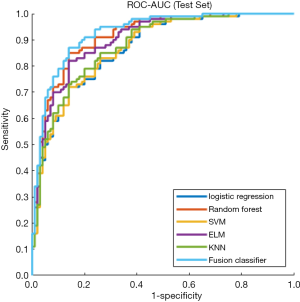
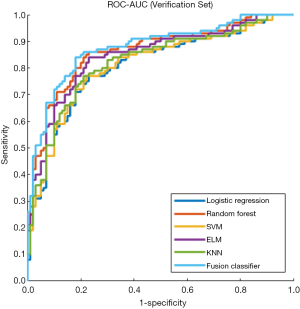
The prediction performance of classifiers
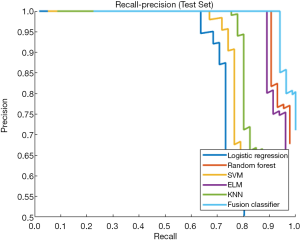
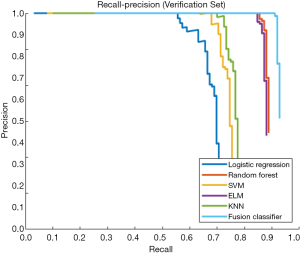
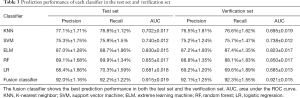
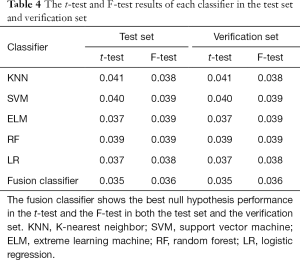
This study employed a simple 10-fold cross-validation method to analyze the performance indicators of the classifiers, and we obtained the ROC curves (Figures 5,6), prediction precision, and recall curves (Figures 7,8) of each classifier. The fusion classifier demonstrated superior prediction performance in the test set (precision =92.0%±1.16%, recall rate =92.2%±1.22%, and AUC =0.915±0.019) and the verification set (precision =92.1%±1.25%, recall rate =92.3%±1.55%, and AUC =0.921±0.015) over any single classifier. After cross-validation, the performance indicators (precision, recall rate, and AUC) all fluctuated within a small range (Table 3), indicating that the fusion algorithm had strong robustness. The t-test and F-test were used to statistically analyze the mean and variance of the prediction performance indicators of the samples, and the results are expressed as P values. The smaller the P value, the greater the probability that the performance indicator of the samples can represent the entire population. The t-test (P=0.035) and F-test (P=0.036) of the fusion classifier showed the optimal null hypothesis performance (Table 4).
Full table
Full table
Discussion
At present, the clinical analysis of pulmonary nodule images is limited to qualitative and preliminary quantitative analysis of the lesions, including observation of the overall and marginal shape of the lesion, the uniformity of internal density, the relationship with the surrounding structure, and a rough measurement of the nodule’s long and short diameters, while no in-depth or detailed analysis of the images is performed. In this study, the benign and malignant nodules overlapped on multiple imaging signs, that is, the so-called same shadow of different diseases. When the volume of the nodule is small, the images would not manifest malignant signs, so it is difficult to characterize the pulmonary nodules simply based on image signs. Radiomics is the application of digital image processing and machine learning techniques to medical image analysis (9). It entails extracting hundreds of quantitative image features from the ROI in the images, followed by screening and analyzing these features to describe the biological characteristics and heterogeneity of the lesions. Therefore, radiomics can identify information that is not visible to the naked eye in conventional imaging images, and it is not limited by lesion size or morphology (10,11).
This study extracted 450 radiomics features from 342 cases of primary pulmonary solid nodules and proposed a multiclassifier fusion method based on radiomics to predict benign and malignant nodules. The main findings include the following. The top 25 features according to weight after feature screening played a major role in the correct classification of the two groups of patients, which included texture features (NGLDM, GLRLM, GLCM, NGTDM, and LBP), wavelet features, and gray-level features. Moreover, the prediction performance of the fusion classifier was better than that of any single classifier.
Gray-level features can quantitatively reflect the amplitude and frequency of pixel-value distribution in the ROI. Kamiya et al. (12) found that compared with benign nodules, malignant nodules showed higher skewness and lower kurtosis in gray-level features. Petkovska et al. (13) reported that comprehensive use of shape, size, and gray-level features could improve the AUC from 0.79 to 0.84 for distinguishing benign from malignant nodules. Chi et al. (14) also found that the skewness and kurtosis demonstrated statistical significance in the identification of benign and malignant nodules in their study of 110 cases of pulmonary solid nodules. Texture features can quantify the subtle differences in image pixel values and their arrangement. Compared with gray-level features, texture features have the advantage of retaining the spatial features of the lesions (15,16). In a study on mediastinal lymph nodes in patients with lung cancer, Bayanati et al. (17) found that entropy, gray-level nonuniformity (GLNU), and running length nonuniformity (RLNU) of the texture features could correctly distinguish the benign and malignant mediastinal lymph nodes in patients with primary lung cancer. In another study on texture features, Chi et al. (18) reported that the contrast, correlation, entropy, and homogeneity had value in the qualitative diagnosis of pulmonary nodules.
Our research not only confirmed that the gray-level features and texture features were important in the classification of pulmonary nodules, it also employed the relief feature selection algorithm to rank the weight of the four categories of features and selected the top 25 features as the input features of the classifiers. The results showed that the gray-level features and texture features had the greatest weights, especially the texture features, which made up the majority of the high-weight features. In contrast to principal component analysis (PCA), the relief algorithm uses the characteristics of pulmonary nodules as the evaluation indices and uses the clustering method for internal calculations. Therefore, this algorithm contains both the external and internal characteristics of pulmonary nodules, while the PCA method only analyzes the internal characteristics of the pulmonary nodule and thus is not representative or conducive to feature selection. Heterogeneity is a recognized feature of malignant tumors, reflecting changes in cell permeability, abnormal angiogenesis, and changes in tissue structure caused by mucus-like changes, necrosis, and fibrosis (19). Therefore, the images of malignant nodules demonstrate an uneven distribution of gray-level, complex texture, and disarranged local texture. To handle this heterogeneity, we introduced the LBP to analyze the local texture features of the images. The LBP features have the advantages of rotation invariance, gray-level invariance, and strong resistance to image noise (20). Feature selection results (LBP feature weights ranked between 25 and 50) showed that LBP was valuable for the qualitative diagnosis of pulmonary nodules. The wavelet features are multiscale features obtained after wavelet transformation of the images, which integrates features at the boundary and vicinity of the lesions and reflects the change rate of the pixel value in the frequency domain (21,22). These features had high weights in our study, indicating that the local texture of malignant nodules was complex, the texture changed quickly, and the lesion boundary was irregular.
In terms of classifier selection, this study proposed a prediction method using multiclassifier fusion (23,24). This method weighted and fused the results of five classic classification methods to obtain the optimal prediction. The method of fusing classifiers first calculates the output weight of each single-classifier prediction to construct an objective function, then calculates the weight corresponding to the single classifier using a Lagrangian and QR decomposition method. Due to the different algorithms and working principles of each single classifier, the sensitivity of different classifiers to different data sets is different, which leads to differences in their predictive performance. The fusion classifier combines the excellent performance of each classifier and has a higher adaptation to the data and better generalization performance. This study used a simple 10-fold cross-validation method to statistically analyze the prediction performance of the fusion classifier and five single classifiers, and the fusion classifier demonstrated the best prediction performance, and the prediction precision, recall rate, and AOC fluctuated within a small range, indicating that the fusion algorithm had high robustness.
To determine whether the prediction performance of the classifier on the experimental samples could be generalized to the whole population, this study used the t-test and F-test to statistically analyze the mean and variance of the prediction performance indicators. The results showed that the performance of this fusion classifier had a probability of 0.965 to represent the mean of the entire population and a probability of 0.964 to represent the variance of the entire population, indicating that this fusion algorithm had strong generalization. Moreover, this fusion classifier integrated the excellent performance of individual classifiers. When the optimal hyperparameters of the classifier and the data set distribution are unknown, a fusion classifier can further simplify the parameter adjustment process.
This study had certain limitations: (I) it was a retrospective study and thus had certain biases. (II) All radiomics features were extracted from manually segmented images. It was difficult to exclude small blood vessels and small bronchi in or around the nodules, which might have affected the precision of the features. (III) The prediction precision of radiomics is affected by the choice of the classifiers, and most of the parameter optimization of the classifiers was done based on experience or experimental adjustment, without the theoretical support of parameter adjustment optimization, which may not guarantee that the parameters reach or approach the optimal performance.
In conclusion, the fusion classifier based on radiomics features can provide a noninvasive, fast, low-cost, and repeatable method to predict benign and malignant pulmonary solid nodules, which will be conducive to clinical treatment.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All experimental procedures were approved by the Medical Ethics Committee of Yinzhou Hospital affiliated with the School of Medicine of Ningbo University, Ningbo, China (ID: 2019-48).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234-48. [Crossref] [PubMed]
- Felix A, Oliveira M, Machado A, et al. Using 3D Texture and Margin Sharpness Features on Classification of Small Pulmonary Nodules. 29th SIBGRAPI Conference on Graphics, Patterns and Images. IEEE, 2016.
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Shen C, Liu Z, Guan M, et al. 2D and 3D CT Radiomics Features Prognostic Performance Comparison in Non-Small Cell Lung Cancer. Transl Oncol 2017;10:886-94. [Crossref] [PubMed]
- Fan L, Fang M, Dong D, et al. Subtype discrimination of lung adenocarcinoma manifesting as ground glass nodule based on radiomics. Chin J Radiol 2017;51:912-7.
- Zhou T, Lu H, Zhang J, et al. Pulmonary Nodule Detection Model Based on SVM and CT Image Feature-Level Fusion with Rough Sets. Biomed Res Int 2016;2016:8052436.
- Li XX, Li B, Tian LF, et al. Automatic benign and malignant classification of pulmonary nodules in thoracic computed tomography based on RF algorithm. IET Image Processing 2018;12:1253-64. [Crossref]
- Peng J, Qi X, Zhang Q, et al. A radiomics nomogram for preoperatively predicting prognosis of patients in hepatocellular carcinoma. Transl Cancer Res 2018;7:936-46. [Crossref]
- Yang M, Ren Y, She Y, et al. Imaging phenotype using radiomics to predict dry pleural dissemination in non-small cell lung cancer. Ann Transl Med 2019;7:259. [Crossref] [PubMed]
- Xu X, Huang L, Chen J, et al. Application of radiomics signature captured from pretreatment thoracic CT to predict brain metastases in stage III/IV ALK-positive non-small cell lung cancer patients. J Thorac Dis 2019;11:4516-28. [Crossref] [PubMed]
- Kamiya A, Murayama S, Kamiya H, et al. Kurtosis and skewness assessments of solid lung nodule density histograms: differentiating malignant from benign nodules on CT. Jpn J Radiol 2014;32:14-21. [Crossref] [PubMed]
- Petkovska I, Shah SK, McNitt-Gray MF, et al. Pulmonary nodule characterization: a comparison of conventional with quantitative and visual semi-quantitative analyses using contrast enhancement maps. Eur J Radiol 2006;59:244-52. [Crossref] [PubMed]
- Chi S. Density histogram analysis of CT scan in the differential diagnosis of solid pulmonary nodule. Radiol Practice 2016;31:866-9.
- Andersen MB, Harders SW, Ganeshan B, et al. CT texture analysis can help differentiate between malignant and benign lymph nodes in the mediastinum in patients suspected for lung cancer. Acta Radiol 2016;57:669-76. [Crossref] [PubMed]
- Ng F, Ganeshan B, Kozarski R, et al. Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology 2013;266:177-84. [Crossref] [PubMed]
- Bayanati H, E, Thornhill R, Souza CA, et al. Quantitative CT texture and shape analysis: can it differentiate benign and malignant mediastinal lymph nodes in patients with primary lung cancer? Eur Radiol 2015;25:480-7. [Crossref] [PubMed]
- Chi S. Differentiation between malignant and benign pulmonary nodules by CT-based texture analysis. J Pract Radiol 2016;32:1789-92.
- Ganeshan B, Goh V, Mandeville HC, et al. Non-small cell lung cancer: histopathologic correlates for texture parameters at CT. Radiology 2013;266:326-36. [Crossref] [PubMed]
- Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Transactions on Pattern Analysis and Machine Intelligence 2002;24:971-87. [Crossref]
- Tu SJ, Wang CW, Pan KT, et al. Localized thin-section CT with radiomics feature extraction and machine learning to classify early-detected pulmonary nodules from lung cancer screening. Phys Med Biol 2018;63:065005. [Crossref] [PubMed]
- Zhang R, Shen J, Wei F, et al. Medical image classification based on multi-scale non-negative sparse coding. Artif Intell Med 2017;83:44-51. [Crossref] [PubMed]
- Ding M, Antani S, Jaeger S, et al. Local-Global Classifier Fusion for Screening Chest Radiographs. SPIE Medical Imaging, 2017.
- Shankar K, Elhoseny M, Lakshmanaprabu SK, et al. Optimal feature level fusion based ANFIS classifier for brain MRI image classification. Concurrency Computat Pract Exper 2018;32:e4887.