
A novel topoisomerase I inhibitor DIA-001 induces DNA damage mediated cell cycle arrest and apoptosis in cancer cell
Introduction
Topoisomerase I (Topo I) is a type IB enzyme with an essential role in controlling the topological structure of DNA during replication, transcription, recombination and repair (1-3). Topo I transiently cut one strand of DNA to form single-strand breaks, allowing supercoiled DNA to relax. For rapid cell division, cancer cells need high Topo I activity to finish the DNA metabolic processes. Due to the specific function of Topo I, inhibiting Topo I can induce DNA double-strand breaks and cellular lethality (4). Therefore, Topo I inhibitors have been developed for cancer therapy (4). However, many Topo I inhibitors, such as camptothecin (CPT) are limited by poor solubility and dose limiting bone marrow toxicity, diarrhea, nausea, and vomiting. The discovery of novel catalytic inhibitor for Topo I is needed.
Topo1 inhibitors are grouped as two types, (I) Topo1 poisons that covalently trap topoisomerases on DNA (5,6). (II) Topo1 catalytic inhibitors, which prevent DNA cleavage (5,6). In our study, the 3Z)-3-[2-(4-Chlorophenyl)-2-oxoethylidene]-1,3-dihydro-2H-indol-2-one (C16H10ClNO2, DIA-001) has been discovered as a novel Topo1 poison and catalytic inhibitor. DIA-001can bind Topo1 to form a Topo1-drug complex that prevents the DNA replication process, induces cell cycle arrest at the G2/M phase and activates the apoptosis pathway. Here, we report the DIA-001 as a novel catalytic Topo1 inhibitor, which binds with the enzyme to decrease the efficiency of supercoiled DNA relaxation and induce DNA damage.
Methods
Cell culture and antibodies
The breast epithelial cell line MCF-10A, mouse muscle myoblast cell line C2C12, human glioma cell lines U251, T98G, LN18, human ovarian cancer cell line OVCAR8, human osteosarcoma cell lines U2OS, human hepatocellular carcinoma cell line HepG2 and human malignant melanoma cell line A375 were purchased from American Type Culture Collection (ATCC, USA). The identities of all these cell lines were confirmed by the medical genome facility at the Mayo Clinic Center (Rochester, MN, USA) using short tandem repeat profiling. Cells were maintained in the appropriate media with 10% FBS.
Antibodies against Chk1, phosphor-Chk1 (Ser345), Chk2, phosphor-Chk2 (Thr68), and cleavage PARP were purchased from Cell Signaling Inc. Anti-γH2AX (05-636) was purchased from Millipore. Anti-β-actin antibodies were purchased from Sigma. Antibodies against Cyclin A, Cyclin B and horseradish peroxidase–conjugated secondary antibodies against mouse (sc-2748) and rabbit (sc-2750) were purchased from Santa Cruz.
MTS and clonogenic assay
For MTS assay, the cells were plated on 96-well plates at densities of 2000 cells per well for U251, OVC8, U2OS, A375, LN18, HepG2 and T98G cell lines respectively in 100 µL/well medium and incubated for 24 hours. Then, fresh medium was added and cells were treated with DIA-001(TimTec) at different concentrations for 72 hours. Cell viability was examined with MTS according to the manufacturer’s instructions (Promega). Briefly, 20 µL of MTS were added to each well 3 hours later. The absorbance at 490 nm was measured with a microplate reader (Tecan Infinite M1000 PRO).
For clonogenic assay, 500 cells were seeded in a 6-well plate for 24 hours and then treated with the indicated concentration of DIA-001. After 10 days, the cells were washed with PBS, fixed with methanol and stained using 0.1% crystal violet and the colony numbers were counted.
Western blotting
Cells were lysed with 0.5% NP40, 50 mMTris, 150 mM NaCl, and 1 mM EDTA (NETN) buffer with 10 mM NaF, 50 mM β-glycerophosphate, and 1 mg/mL each of aprotinin and pepstatin A. Proteins were separated by SDS-PAGE, transferred onto PVDF membrane and probed using appropriate primary and secondary antibodies. Finally, proteins were detected using ECL Western Blotting Detection Reagents (ThermoFisher).
Immunofluorescence for nuclear foci
Cells were seeded on a coverslips. After treatment, cells were washed with PBS and fixed with 3% paraformaldehyde for 15 minutes, permeabilized with 0.5% Triton-X in PBS for 5 minutes and following with blocking by 5% goat serum for 1 hour at room temperature. Then, cells were incubated with primary antibody at 4 °C overnight, followed by incubation with Alexa Fluor 594 or Alexa Fluor 488 conjugated secondary antibodies at 37 °C for 20 minutes. After washed with PBS, cell nuclei were counterstained with DAPI. The signals were examined by confocal microscopy.
Cell cycle analysis
Cells were collected and fixed with 70% ethanol at −20 °C overnight, then washed with PBS twice. Cells were stained with propidium iodide (PI) containing RNase for 30 minutes in the dark, then analyzed for cell-cycle profile by FACS (Beckman Coulter). Data were analyzed using ModFit LT software.
TOPO I and II mediated DNA relaxation assays
The effects of DIA-001 on Topo I and Topo II activities were examined by the conversion of supercoiled pBR322 DNA to its relaxed form using topoisomerase I and II drug screening kits (Topogen) according to the manufacturer’s protocol. Briefly, The Topo I activity was studied in a 20 µL reaction system including 1 µL human Topo I, variable volume of DIA-001 (final concentrations: 10, 20 µM) or CPT (50 µM), 4 µL 5× complete assay buffer, 1 µL pBR322 DNA, and variable volume H2O (to a final volume of 20 µL). These mixtures were incubated at 37 °C for 30 min and the reaction stopped using 2 µL 10% SDS. The samples were run on a 1% agarose gel with 0.5 µg/mL ethidium bromide at 50 V. The gel was destained in water before being photographed under UV-light. The known Top I poisons CPT was used as the positive control. The effect on Topo II activity by DIA-001 was performed using a similar procedure of Topo I. The known Topo II inhibitor etoposide was used as the positive control.
Statistics
All the data are presented as mean ± SD. Statistical analysis was performed with Graphpad8 software (GraphPad Inc. USA). Two tailed Student’s t-test was used to evaluate statistical significance between groups. P<0.05 was considered significant difference. In the figures, the statistical significance is showed by: n.s. no significance, *P <0.05; **P<0.01, ***P<0.001.
Results
DIA-001 inhibits the proliferation of cancer cells
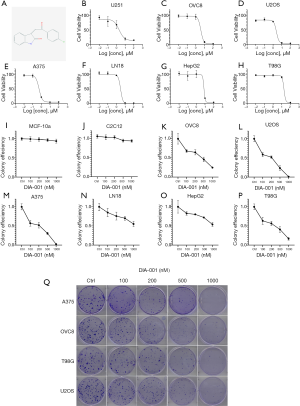
To evaluate the effect of DIA-001 on proliferation inhibition on different cancer cells, the MTS assay and colony formation assay were performed. The cytotoxicity was determined by IC50 calculations for treatment of U251, OVC8, U2OS, A375, LN18, HepG2 and T98G cells by DIA-001 were 1.987, 3.782, 2.425, 0.5399, 3.031, 8.279 and 14.20 µM respectively (Figure 1A,B,C,D,E,F,G,H). Colony formation results showed the same growth inhibition on different cancer cell lines of DIA-001, while this compound did not have cytotoxic effect for normal cells MCF-10A and C2C12 in concentrations up to 1 µM (Figure 1I,J,K,L,M,N,O,P). Collectively, these results indicate that DIA-001 has anti-tumor effect for different cancer cell lines.
DIA-001 Induces DNA damage
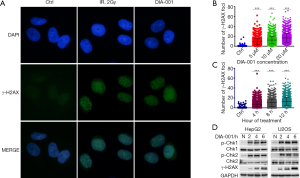
DNA damage is a common reason for inhibition of proliferation and cytotoxicity (7,8). We investigated whether DIA-001 would cause DNA damage in U2OS cell through by examine the phosphorylation of the checkpoint kinase 1, 2 (p-Chk1/2), and H2AX (γH2AX). γH2AX staining, a marker of DNA damage, was used to assess the level of DNA damage (9-11). As shown in Figure 2A,B,C, DIA-001 treatment induced γH2AX foci formation and the number of γH2AX foci increased in a time and dose-dependent manner. Furthermore, western blot analysis showed that the DIA-001 treatment induced H2AX and Chk1/2 phosphorylation levels (Figure 2D). Collectively, these results indicate that DIA-001 markedly induced DNA damage in cancer cell.
DIA-001 induces G2/M arrest and apoptosis in cancer cells
The DNA damage checkpoint is a mechanism that triggers cell cycle arrest to allow enough time for repair (12-14). To uncover the mechanisms of DIA-001 in inhibiting cell proliferation, we studied the cell cycle profiles of U2OS cells after DIA-001 treatment. The result shows that after 24-hour of treatment with 10 and 20 µM DIA-001, U2OS cells arrested at the G2/M phase (Figure 3A,B). Western blot analysis of the cell cycle regulators cyclin A and B in U2OS cell treated with 10 and 20 µM DIA-001 for 24 hours also showed that DIA-001 induced an accumulation of cyclin A and a slight increase of cyclin B, supporting the G2/M arrest (15-17). We also examined whether DIA-001 could induce apoptosis. Cleavage of PARP serves as a marker for cells undergoing apoptosis (18-20). We found that PARP cleavage increased in U2OS cell at 24-hour of DIA-001 treatment (Figure 3C). Our results suggest that DIA-001 induces cell cycle arrest and apoptosis in cancer cell lines.

DIA-001 causes accumulation of Topo1cc
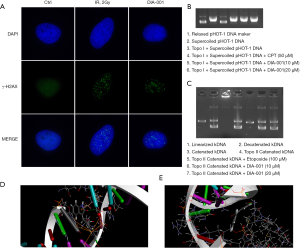
We next explored how DIA-001 causes DNA damage. Interestingly, we found that Topoisomerase might be a target. As shown in Figure 4A, we found the accumulation of Topo1 cleavage complexes (Top1cc) in U2OS cells 4 hours after DIA-001 treatment (CPT treatment was used as a positive control). We hypothesized that DIA-001 might be a Top1 inhibitor. To determine the mechanism of DIA-001 induced Top1cc accumulation, we used the in vitro enzyme assays to test for topoisomerase activity. As shown in Figure 4B, supercoiled pHOT1 DNA cannot be relaxed by topo I with DIA-001 at 10 and 20 µM (lane 5 and 6). To investigate whether the cytotoxicity of DIA-001 is specific for topo I, we used the kDNA decatentation assay, which measures topo II activity in vitro. The kinetoplast DNA (kDNA) is an ideal substrate for topoisomerase II assays because it is specific for the TopoII reaction mechanisms. As shown in Figure 4C, kDNA was decatentated in the presence of topo II (lane 4) and in the presence of DIA-001 (lane 6–7). In contrast, the topo II inhibitor, etoposide inhibited the decantenation (lane 5). To further elucidate the mechanism of DIA-001 inhibition through formation of a drug-enzyme-DNA complex, we analyzed the predicted topoisomerase enzyme binding site of DIA-001. As shown in Figure 4D,E, the 3-D structure of the Topo I and II shows DIA-001 fits into the binding site of Topo I rather than the Topo II.
Discussion
DNA topoisomerases were recognized as promising targets in cancer, considering its pivotal role in essential biological process (4,21-23). Currently approved topoisomerase inhibition therapies are directed toward blocking the topoisomerase I and topoisomerase II activities that participate in the winding or unwinding of DNA. Topo I inhibitors, such as CPT, are currently used in the treatment of cancer (23,24) and always combine with radiotherapy that is standard treatment for cancer patients (25,26). It has remarkable anticancer activity in the clinic, but also has some limitations with different cancer types. In vitro Topo I assay and molecular modelling study show the specific complex-based pharmacophores. The docked model of DIA-001 with Top I-DNA complex, which is hydrogen-bond formed between the ligand and residues are similar with CPT (23). The combination mode between DIA-001 and Top I-DNA complex induced trapping of Top I by DIA-001 generous Top Icc. Finally, the two ends cannot be relegated induced DNA replication fork stop activated DNA damage pathway and apoptosis. Here we identified DIA-001, a novel Topo1 inhibitor, that is effective in the treatment of various cancers such as small cell lung carcinoma, oral squamous cell carcinoma and glioblastoma in vitro, but is comparatively not toxic for normal cells.
Based on the previous research, the anticancer effect of topoisomerase inhibitor is dependent on the pro-apoptotic and cell cycle arrest effect. Consistent with this, using U2OS cell line treated with DIA-001, we showed that PARP cleavage was enhanced. We also show that cyclin A levels increase after treatment with DIA-001 but cyclin B level increase only slightly suggesting cell cycle progression through the S/G2 transition but arrested at the G2/M phase (15-17). Together, the data from our study indicates that treatment with DIA-001 may induce cell cycle arrest in G2 phase and apoptosis in cancer cells.
In this study, we identified a new Topo1 inhibitor for cancer patient chemotherapy that induces DNA replication stress by accumulation of the Topo1cc on DNA and discuss the potential of using replication stress to promote the activation of DNA damage repair pathway, cell cycle arrest and apoptosis.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (91749115 to J Yuan, 81572770 to K Luo, 81972363 to S Zhao).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Nitiss JL. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim Biophys Acta 1998;1400:63-81. [Crossref] [PubMed]
- Wang JC. DNA topoisomerases. Annu Rev Biochem 1996;65:635-92. [Crossref] [PubMed]
- Leppard JB, Champoux JJ. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma 2005;114:75-85. [Crossref] [PubMed]
- Delgado JL, Hsieh CM, Chan NL, et al. Topoisomerases as anticancer targets. Biochem J 2018;475:373-98. [Crossref] [PubMed]
- Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem Rev 2009;109:2894-902. [Crossref] [PubMed]
- Xu Y, Her C. Inhibition of Topoisomerase (DNA) I (TOP1): DNA Damage Repair and Anticancer Therapy. Biomolecules 2015;5:1652-70. [Crossref] [PubMed]
- Dusre L, Covey JM, Collins C, et al. DNA damage, cytotoxicity and free radical formation by mitomycin C in human cells. Chem Biol Interact 1989;71:63-78. [Crossref] [PubMed]
- Bandi S, Viswanathan P, Gupta S. Evaluation of cytotoxicity and DNA damage response with analysis of intracellular ATM signaling pathways. Assay Drug Dev Technol 2014;12:272-81. [Crossref] [PubMed]
- Bonner WM, Redon CE, Dickey JS, et al. GammaH2AX and cancer. Nat Rev Cancer 2008;8:957-67. [Crossref] [PubMed]
- Tu WZ, Li B, Huang B, et al. γH2AX foci formation in the absence of DNA damage: mitotic H2AX phosphorylation is mediated by the DNA-PKcs/CHK2 pathway. FEBS Lett 2013;587:3437-43. [Crossref] [PubMed]
- Podhorecka M, Skladanowski A, Bozko P. H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. J Nucleic Acids 2010;2010. doi: 10.4061/2010/920161. [Crossref]
- Chao HX, Poovey CE, Privette AA, et al. Orchestration of DNA Damage Checkpoint Dynamics across the Human Cell Cycle. Cell Syst 2017;5:445-59.e5. [Crossref] [PubMed]
- Santivasi WL, Xia F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid Redox Signal 2014;21:251-9. [Crossref] [PubMed]
- Sakaue-Sawano A, Kobayashi T, Ohtawa K, et al. Drug-induced cell cycle modulation leading to cell-cycle arrest, nuclear mis-segregation, or endoreplication. BMC Cell Biol 2011;12:2. [Crossref] [PubMed]
- Shin DY, Sung Kang H, Kim GY, et al. Decitabine, a DNA methyltransferases inhibitor, induces cell cycle arrest at G2/M phase through p53-independent pathway in human cancer cells. Biomed Pharmacother 2013;67:305-11. [Crossref] [PubMed]
- Hsiao YP, Tsai CH, Wu PP, et al. Cantharidin induces G2/M phase arrest by inhibition of Cdc25c and Cyclin A and triggers apoptosis through reactive oxygen species and the mitochondriadependent pathways of A375.S2 human melanoma cells. Int J Oncol 2014;45:2393-402. [Crossref] [PubMed]
- Hiraoka D, Aono R, Hanada S, et al. Two new competing pathways establish the threshold for cyclin-B-Cdk1 activation at the meiotic G2/M transition. J Cell Sci 2016;129:3153-66. [Crossref] [PubMed]
- McGowan AJ, Ruiz-Ruiz MC, Gorman AM, et al. Reactive oxygen intermediate(s) (ROI): common mediator(s) of poly(ADP-ribose)polymerase (PARP) cleavage and apoptosis. FEBS Lett 1996;392:299-303. [Crossref] [PubMed]
- Oliver FJ, de la Rubia G, Rolli V, et al. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem 1998;273:33533-9. [Crossref] [PubMed]
- Aslan Koşar P, Tuncer H, Cihangir Uğuz A, et al. The efficiency of Poly(ADP-Ribose) Polymerase (PARP) cleavage on detection of apoptosis in an experimental model of testicular torsion. Int J Exp Pathol 2015;96:294-300. [Crossref] [PubMed]
- Jain CK, Majumder HK, Roychoudhury S. Natural Compounds as Anticancer Agents Targeting DNA Topoisomerases. Curr Genomics 2017;18:75-92. [Crossref] [PubMed]
- Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem Biol 2013;8:82-95. [Crossref] [PubMed]
- Li F, Jiang T, Li Q, et al. Camptothecin (CPT) and its derivatives are known to target topoisomerase I (Top1) as their mechanism of action: did we miss something in CPT analogue molecular targets for treating human disease such as cancer? Am J Cancer Res 2017;7:2350-94. [PubMed]
- Puddu F, Salguero I, Herzog M, et al. Chromatin determinants impart camptothecin sensitivity. EMBO Rep 2017;18:1000-12. [Crossref] [PubMed]
- Yan Y, Li Z, Zeng S, et al. FGFR2-mediated phosphorylation of PTEN at tyrosine 240 contributes to the radioresistance of glioma. J Cell Commun Signal 2019;13:279-80. [Crossref] [PubMed]
- Xu Z, Yan Y, Xiao L, et al. Radiosensitizing effect of diosmetin on radioresistant lung cancer cells via Akt signaling pathway. PLoS One 2017;12:e0175977. [Crossref] [PubMed]


