
Tumor slice culture as a biologic surrogate of human cancer
Introduction
In the current era of molecular oncology, patient-derived models that can accurately inform clinical decisions are urgently needed. Cancer is a diverse collection of diseases with enormous biologic and genomic heterogeneity, thus precision medicine, i.e., matching the right drugs with the right patients, has been an ongoing challenge. Advances in the molecular profiling of tumors through next-generation sequencing have led to new treatment paradigms with astonishing results for select patients, but it is estimated that fewer than 5% of patients with advanced cancers currently benefit from this approach (1). Many reasons contribute to this lackluster result, including a shortage of functional assays, the absence of effective therapies targeting common cancer genes such as RAS and p53, and the emergence of resistance stemming from functional compensation by other genes (2-4). Further, a single mutation may play different roles in different tumors, as highlighted by the variable response of BRAF (V600E)-associated non-melanoma cancers to the BRAF inhibitor vemurafenib (5). At present, therapeutic decisions are based on a limited set of ‘actionable’ genomic data, without a full understanding of its biologic significance. Thus, a lack of individualized functional assays is a major shortcoming in the current practice of precision oncology.
Human cancer models have become more prevalent with the increasing availability of organoid technology and in vivo patient-derived xenograft (PDX) mouse models (6,7). While these methods have yielded valuable biologic insights, one shared pitfall of these techniques is the inability to maintain the original tumor microenvironment (TME). Organotypic tissue slice cultures (TSCs) have been used for decades to study the physiologic properties of intact tissues, such as those of the central nervous system (8). Recent studies using TSCs as an ex-vivo model to predict drug sensitivity in cancer therapy have led to renewed interest in this platform, with investigators demonstrating preservation of the TME, molecular signaling, and treatment responses (9-12). Recently, we reported that slices from human pancreatic cancers remain viable in vitro and retain their TME, including the local immune milieu, thus allowing investigations of tumor immunology (13). Widespread application of TSCs in personalized oncology has lagged due to the absence of standardized methods for comparison between samples. Furthermore, there has been a paucity of evidence demonstrating that organotypic cultures reflect clinical characteristics of human cancers. Here we report a simple protocol that can be applied to diverse solid human gastrointestinal malignancies, and demonstrate the biologic relevance of TSCs by comparing in vitro growth properties with clinical and pathologic parameters. Using standardized readouts, this system allows us to compare and characterize inter- and intra-tumoral heterogeneity that correlates with the biologic state of the disease.
Methods
Subjects and clinical data
Patients undergoing liver resection for primary or metastatic tumors were consented for tissue donation under IRB-approved protocols (University of Washington IRB #1852 and #1666). Fresh biospecimens were collected from a total of 108 patients over a two-year period, along with relevant de-identified clinical data.
Sample preparation
Sterile specimens from hepatectomies were processed immediately at the completion of resection under the direction of the surgeons (JO Park, RS Yeung) in consultation with Pathology staff (M Yeh). To maintain maximal sterility of the specimens, the latter were placed in a sterile field within the operating room. Following the directives of the pathology representative, ‘to-be-discarded’ tissues were procured in a sterile environment, usually within 10 minutes of the resection.
Tissue procurement
In order to establish a reproducible protocol that allows for comparison between samples, we aimed to generate tissue slices of a fixed volume. Six-mm tissue cores were procured from specimens using a sterile biopsy punch (Integra Miltex, York, PA), and immediately placed in cold Belzer-UW solution (Bridge to Life Ltd., Columbia, SC) prior to transfer to research laboratories. We found that 6-mm cores are optimal for tumors >2 cm without interfering with clinical diagnosis. In our hands, smaller cores down to 4-mm worked equally well although the signals from the MTS assay were proportionally reduced.
Precision-cut slices
Please refer to Supplemental Methods for step-by-step protocol. Briefly, 250 µm thick tumor slices were cut using a Leica VT 1200S vibrating microtome (Leica Biosystems Nussloch GmbH, Germany) with the following settings: amplitude 2–3 mm and speed 0.5–1.5 mm/s, depending on tissue consistency. Tissue slices were washed with Williams’ Media E (Gibco, Grand Island, NY, USA), placed on Millicell Cell Culture Inserts (0.4 µm PTFE, EMD Millipore, Burlington, MA) in 24-well cell culture plates containing growth media, and incubated at 37 °C in 5% CO2 on a lab rocker set at 20 rotations/min.
Culture conditions
Tumor slices were cultured in Williams’ Media E containing nicotinamide (12 mmol/L), L-ascorbic acid 2-phosphate (50 mg/mL), D-(+)-and Glucose (5 mg/mL) (Sigma-Aldrich, St. Louis, MO, USA); Sodium Bicarbonate (2.5%), HEPES (20 mmol/L), Sodium Pyruvate (1 mmol/L), Glutamax (1%), and Penicillin Streptomycin (0.4%) (Gibco, Grand Island, NY, USA); ITS + Premix (1%), Human EGF (20 ng/mL) (BD Biosciences, San Jose, CA, USA). Media were changed one day after slicing and then every 2–3 days.
MTS assay
Cell viability was assessed using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium; MTS) (Promega, Fitchburg, WI, USA). Slices were incubated with MTS reagent while rocking at 37 °C for 2 or 3 hours. Media were transferred to a 96 well plate; absorbance was read at 490 nm and normalized to blanks (wells containing media and MTS without tissue slices). After MTS measurements, slices were either fixed in 10% formalin for 24 hours at 4 °C or placed back in their original cell inserts for continued culturing.
Histology and immunohistochemistry
Fixed slices were embedded in paraffin, and 4µm thick sections were cut and placed on glass slides. Slides were stained with hematoxylin and eosin or processed for Ki-67 immunohistochemistry (IHC) as described (14). To determine the Ki-67 positive fraction, the number of stained cells in at least 10 high power fields was counted.
Tumor cell transcriptomics
Tumor slices were dissociated using the gentleMACS system (Miltenyi Biotec) and loaded on a Chromium instrument (10X Genomics) for single-cell RNA processing and 150 cycles of NextSeq500 (Illumina) sequencing. Reads were aligned to the human genome (GRCh38) and quantified using CellRanger v2 software. Tumor and non-tumor cells were computationally segregated by K-means clustering. Average CEACAM5, KRT20, and GAPDH expression levels were calculated for the EPCAM-expressing tumor cell clusters.
Statistical analyses
R statistical software was used to develop a predictive model for high MTS values based on univariate and multivariate linear regression. Clinical covariates included age, sex, body mass index (BMI), history of smoking, any prior cancer treatments (surgical resection, radiofrequency ablation (RFA), chemotherapy, etc.), number of prior chemotherapy regimens, maximum liver tumor and primary tumor size of non-liver tumors, known metastatic disease, and number of tumors. Pathologic covariates included histology, maximum focus size and percent of viable/necrotic tissue in resected specimens, tumor regression grade (TRG) score, tissue fibrosis, tissue differentiation, maximum mitotic activity per high power field, and presence of nodal disease. Data were analyzed as continuous or discrete numeric variables, or as categorical or binary variables. Results for univariate and multivariate analyses are reported as slope coefficients for covariates of interest with 95% confidence intervals calculated from robust standard errors, P values for coefficients, and R2 values for each model. For the leave one out cross validation (LOOCV) analysis, results presented for comparison are the covariates included in each model, root-mean-squared errors (RMSE), and R2 values to compare predictive accuracy of the models.
Results
Standardized assessment of human cancer tissue slice cultures
We set out to standardize our TSC platform, which allowed us to compare results between different tumor samples. As a quantitative readout, we used a colorimetric MTS assay to measure metabolic activity as our primary endpoint (15). This assay has the key advantage of low toxicity of MTS reagent, enabling multiple assessments of the same sample over time. In our experience, MTS absorbance remains stable over at least 3 repeated assays on any one sample (data not shown). Next, we considered a number of methods to standardize MTS absorbance across different samples, including normalizing data to the wet weight of the slices, tissue volume, or total protein content. The latter method requires destruction of the sample, which would eliminate our ability to perform longitudinal studies such as drug sensitivity testing. Instead, we standardized the volume of tissue in our cultures using a precision-cutting vibratome (Leica) to generate 250 µm thick slices from 6-mm cores, resulting in thin cylinders with a fixed volume of approximately 7 mm3 (Figure 1A). Baseline MTS measurements from consecutive slices from the same core were determined with and without normalization to the wet weights of individual slices. Figure 1B shows the results from two colorectal carcinoma (CRC) TSCs, highlighting significant variability in the wet weights of the slices, while baseline MTS absorbances were highly homogeneous. In our hands, we found greater variability when the colorimetric absorbance was normalized to the wet weights of each slice. We attribute these results to our inability to precisely control the fluid or water content of slices during the weighing process while maintaining tissue viability, and postulate that this water content does not affect MTS values. Therefore, standardizing the slice dimensions without normalization had significantly less variance (Figure 1B table). We further confirmed that consecutive precision-cut slices from the same core of tumors of various histologies have nearly identical baseline MTS absorbance, obviating the need for additional normalization (Figure 1C). The simple step of standardizing the dimensions of the tumor slices allowed us to obtain highly reproducible assessment of the global ‘health’ of individual slices at baseline, thus enabling us to compare results among samples.
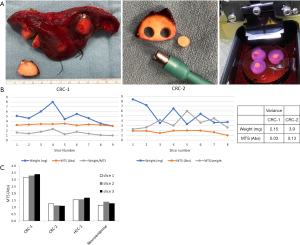
Standardized assessment of tumor slice cultures (TSCs) by precision cutting. (A) Workflow of the TSC platform from surgical specimen to slices. Left, representative tumor ‘wedge’ removed from surgical specimen; middle, 6-mm core from a punch biopsy; right, agarose-embedded cores set in Vibratome for slicing. Refer to Supplemental Methods for details; (B) comparison of baseline MTS absorbance with and without normalization to wet weight of slices from 2 cases of metastatic colorectal carcinoma (CRC). The table shows the variance of weight and absorbance measurements; (C) baseline MTS absorbance of 3 consecutive TSCs each from 4 different tumors without normalization to wet weight. HCC, hepatocellular carcinoma.
Culture viability and integrity
Next, we examined the viability of TSCs over time. In the majority of cases, MTS measurements did not change over the first week in culture, as illustrated by the four independent tumors shown in Figure 2A. In further support of in vitro maintenance of growth, corresponding Ki-67+ fractions in each of these TSCs remained stable over the 7-day period (Figure 2B). Figure 2C shows tumor histology and Ki-67 IHC in CRC, hepatocellular carcinoma (HCC), and intrahepatic cholangiocarcinoma (ICC) TSCs on days 0 and 7, demonstrating that tumor architecture is preserved over time in TSCs, as are their proliferative activities.
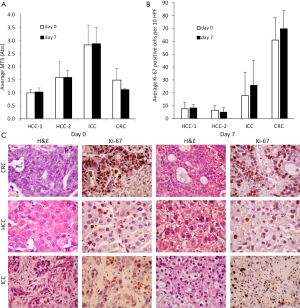
In our experience with 108 consecutive tumors, we were able to generate TSCs in 89% of cases, thus making this technique highly reliable and efficient in creating patient-specific cancer models for short-term investigation. Of the different tumor histologies from which we have generated TSCs, CRCs best maintain their ‘health’ in vitro, with a survival rate of 98%, while HCCs and ICCs show greater decline in their viability over the course of a week (65% and 71% survival, respectively). In an attempt to optimize conditions for HCC slices, we varied levels of serum, glucose, growth factors, and matrix in our cultures, but did not observe a consistent improvement in TSC survival over a two-week period with any of these changes (data not shown). As a result, we used uniform media and growth conditions for all tumor samples going forward (i.e., after our first 30 cases). Our current protocol is detailed in the Supplemental File.
Correlation of TSC viability with clinical and pathologic parameters
We tabulated baseline MTS absorbances from 53 consecutive samples representing diverse tumor types following the standardization of our protocol; the clinical characteristics of the study cohort are summarized in Table S1. Figure 3 shows the mean baseline MTS absorbance from a minimum of three TSCs derived from 51 of these 53 cases; two cases in this initial cohort failed to cut due to poor tumor consistency. One can readily appreciate the wide range of baseline MTS values (i.e., 0.076 to 3.396), indicating the tremendous variability in growth and proliferative activity among different tumors at the time of resection.
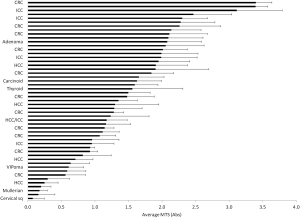
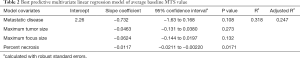
We postulated that baseline MTS values reflect the variable biologic activities of the in vivo state, and may correlate with clinical and pathologic features. Univariate analyses based on the clinical and pathologic parameters shown in Table 1 identified three factors that correlated with in vitro viability scores (MTS values): (I) maximum tumor size on preoperative imaging (slope coefficient b =−0.083, P=0.02); (II) “other” histologic cancer type (b =−0.70, P=0.02); and 3) percent tumor necrosis on histologic examination (b =−0.012, P=0.005). Baseline MTS activities did not meaningfully differ between CRC, HCC, or ICC, nor did it correlate with age, sex, history of smoking, prior cancer treatment, number of neoadjuvant chemotherapy regimens, tumor regression grade, tissue fibrosis, tissue differentiation, mitotic activity, or nodal disease. Primary tumor size on preoperative imaging (b =−0.078, P=0.06), metastatic disease (b =−0.56, P=0.10), maximum viable focus size (b =−0.064, P=0.07), and percent tumor viability (b =0.006, P=0.10) trended toward associations with in vitro viability, but did not reach statistical significance on univariate analysis.
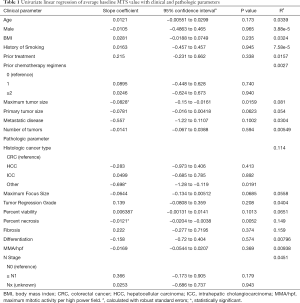
Full table
A multivariate linear regression model chosen a priori, controlling for histologic cancer type, metastatic disease, maximum tumor size, maximum focus size, and percent necrosis, found that lower mean percent necrosis remained associated with higher baseline mean MTS (b =−0.012, 95% CI: −0.0211, −0.00273, P=0.01). Other covariates in the multivariate model did not show a significant association with mean MTS, though histologic cancer type of “other” still showed a trend of lower mean MTS compared to CRC as a reference (b =−0.471, P=0.08).
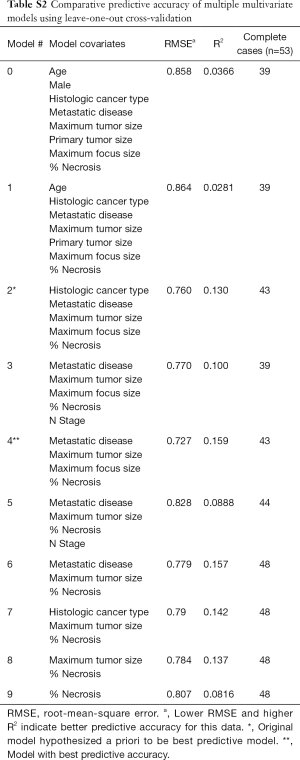
Comparing this model with several others for internal validity, we found a consistent association between mean percent necrosis and mean baseline MTS regardless of the number of other covariates included in the model. Using LOOCV with complete cases as described in methods, we determined that a model including metastatic disease, maximum tumor size, maximum viable focus size, and percent necrosis (Table 2) had the best predictive accuracy for in vitro viability, given it is lowest comparative RMSE and highest comparative R2 (Model #4 in Table S2). Together, these findings indicate that baseline MTS absorbance in TSCs mirrors tumor viability at the time of resection across all tumor types.
Full table
Full table
Assessment of tumor heterogeneity
While there is clear evidence that genomic and transcriptomic heterogeneity exist within different parts of a given tumor, less is known about intra-tumoral variability in terms of metabolic activity. Our ability to detect metabolic differences among tumors (see Figure 3) suggested that our TSC method could also identify metabolic heterogeneity within tumors. We examined heterogeneity within tumors at two levels: regional differences that span several centimeters within a larger tumor, and local heterogeneity within a few millimeters. Figure 4A shows examples of baseline MTS values from separate tumor cores separated by at least 1 cm obtained from tumors that were >3 cm in diameter, i.e., regional differences within tumors. Given that the central portion of a large solid tumor is often necrotic, we purposely took samples from the periphery of tumors, where tissue is expected to be more viable. Nonetheless, we observed significant differences in baseline MTS values from separate sites within each tumor in 4 out of 5 cases (Figure 4A). We found that the variation in baseline metabolic activity correlates with the distribution of tumor vs. stromal components represented by each slice. Figure 4B illustrates the histology of the 3 sites from CRC-1 shown in Figure 4A. Of the 3 areas, Site 2 has extensive collagen deposition with only small areas of necrotic tumor cells, whereas Sites 1 and 3 contain significantly more viable tumor cells. Correspondingly, Site 2 had the lowest baseline MTS absorbance. On closer inspection, tumor cells occupy a significantly larger area of Site 1 compared to Site 3 despite similar MTS absorbance. We thus hypothesized that tumor cells in site 3 were more mitotically and/or metabolically active than at the other two sites. Ki-67 IHC demonstrated a significantly higher fraction of Ki-67+ tumor cells in site 3 compared to site 1 (82% vs. 21%; Figure 4B). These data suggest that both tumor cell density and their proliferative activities influence baseline MTS values in CRC metastases.
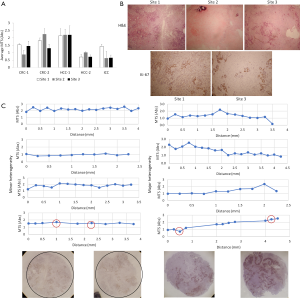
To examine local heterogeneity within a span of millimeters, we analyzed consecutive tumor slices over the length of individual tissue cores (~1 cm). Of the eight samples analyzed in this fashion, four had similar MTS values (i.e., minor heterogeneity is defined as variability within 1 unit of absorbance) across the length of the cores, while the other 4 showed significant heterogeneity over similar distances (i.e., >1 unit of drift in absorbance; Figure 4C). Histologic examination revealed a correlation between MTS absorbance and the proportion of viable cancer cells within each slice (Figure 4C, bottom). Thus, within the CRC tumor microenvironment, tumor cells (as opposed to stromal cells) are the major contributor of metabolic activity in slice cultures.
Tumor CEA correlates with clinical response to chemotherapy
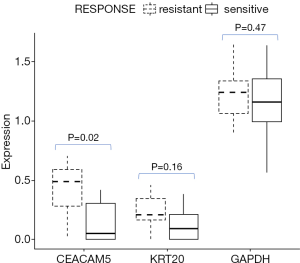
Baseline serum carcinoembryonic antigen (CEA) has been shown to be an independent poor prognostic factor in advanced CRC (16). Here, we measured mRNA expression of CEACAM genes from CRC tumor slices in culture for 72 hrs based on RNAseq. Figure 5 illustrates that tissue CEACAM5 (gene encoding CEA) mRNA levels were significant higher in tumor slices from patients who were deemed clinically chemo-resistant. In other words, CEACAM expression inversely correlated with clinical response. This was not due to differences in the viability of the slices between chemo-resistant and chemo-sensitive tumors, given comparable expression of GAPDH and cytokeratin 20 (KRT20) genes.
Discussion
In this study we describe a standardized method to evaluate human-derived TSCs in vitro across a broad spectrum of primary and metastatic liver cancers. Specifically, we developed a simple and reproducible protocol that maintains tumor viability for days, and allows for comparison among fixed-volume samples generated by precision cutting with a vibratome. While previous studies have emphasized the utility of organotypic slice cultures to test drug sensitivity in individual cancers, few studies have compared biologic endpoints among different tumors. Here, we show that baseline MTS absorbance of fixed volume slices provides a quantitative measure of the metabolic activity that accurately reflects disease viability and biologic status of the tumor at the time of resection. Applying the MTS assay across different TSC samples provides a simple way to compare cancer metabolic activity and viability in a cohort of patients.
In our experience of over 100 cases, we have optimized and standardized our protocol for multiple types of primary and secondary liver tumors. We have learned that consecutive slices of fixed dimensions have nearly identical metabolic/growth activity, and thus serve as biologic replicates in experiments. By the same token, we can exploit TSCs to evaluate tumor heterogeneity in human cancers. Tumor tissues that are separated by even a few millimeters can exhibit significant changes in MTS values, reflecting underlying differences in the distribution of tumor cells within a cancer. In our tumor samples from liver metastases, the majority of patients had received pre-operative systemic therapy. The correlation between baseline MTS absorbance and the amount of histologically necrotic tumor allows us to quantitatively gauge the effectiveness of neoadjuvant therapy at the time of resection, independent of standard pathologic evaluation. For CRCs, we also found that tissue CEA mRNA levels negatively correlated with clinical response to chemotherapies.
A key advantage of using TSC as a model of human cancer is the ease and efficiency in generating an in vitro system that preserves the tumor microenvironment. We have exploited this feature in addressing the immune landscape in human malignancies (13,17) that is not easily reproduced in other systems including cell lines, organoids, and PDX mice. There is no lag time associated with TSC; slices can be put to use almost immediately upon culture and without concerns for selection pressure. With a success rate of nearly 90% in establishing TSC in our sample set, the model provides a simple method to study human cancers while preserving the original spatial architecture and elements of the TME.
There are a few important limitations in using the TSC platform. First and foremost is that the lifespan of tumor slices is finite and non-renewable, thus long-term or repeat experiments are limited. Efforts are currently underway to optimize cryopreservation methods such that the slices can be studied at any time. Not all tumors behave the same; in our experience, metastatic colorectal metastases are the most hardy to grow in vitro unless they have undergone extensive necrosis from prior chemotherapy. On the other hand, tumors such as hepatocellular carcinoma are more variable in their ability to remain viable long-term in culture. For any particular sample, there remains many conditions that can be fine-tuned for optimal growth including, but not limited to, components of the media, thickness of the slice, ambient oxygen level, and the extent of physical motion. Our protocol provides a starting point for anyone who wants to explore this method in their studies. Access to fresh, sterile tumor specimens is critical to creating TSCs, and at our Institution, this effort is led by surgeons who, in collaboration with pathologists, have developed a highly efficient practice of procuring samples without compromising clinical care. In the future, we anticipate modification of our technique to adopt core-needle biopsies, which will expand the clinical applications greatly in the field of precision oncology by bridging ‘omics’ data with functional biologic assays.
In summary, our study highlights a simple, standardized method to generate TSCs from solid tumors using a fixed volume of tissue, which allows for sample comparison. Besides the ability to test drug sensitivity, properties inherent in the tumor slices provide biologic information that are clinically relevant. We show that TSCs provide a functional platform that is useful in detecting inter- and intra-tumor heterogeneity, a feature that needs to be taken into consideration in order to optimize treatment recommendations and improve outcome for individual cancers.
Supplementary
Supplemental methods
Reagents
Belzer UW Cold Storage Solution (Bridge to Life)
5 mL Eppendorf tubes (cat. no. 0030119487)
48-well cell culture plates (Corning, C3548)
24-well cell culture plates (Corning, C3527)
96-well assay plate—Corning Costar Assay Plate, clear, flat bottom, 9017
Millipore Cell Culture Inserts (PTFE 0.4µM pore) PICM01250 Millipore Sigma
#10 Scalpels (Exel International, 29550)
Sterile gauze (Covidien, Curity 3157)
Sterile petri dish (Fisher Scientific, FB0875713)
CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS), (Promega,G3580)
UltraPure Low Melting Point Agarose (cat. no. 16520100)
DPBS (Gibco – 14190144)
Williams medium E (Gibco-Invitrogen cat# 12551-032)
L-Glutamine (Gibco-Invitrogen cat#25030-081)
Nicotinamide (Sigma N-0636)
Asorbic acid (Sigma cat# A8960)
Sodium Bicarbonate (Gibco-Invitrogen cat#15630-080)
D-Glucose (Sigma cat# G5767-500G)
Sodium Pyruvate (Gibco-Invitrogen cat#11360-070)
HEPES (Gibco-Invitrogen cat#15630080)
ITS + Premix (BD Biosciences cat#354352)
Penicillin-Streptomycin (Gibco-Invitrogen cat# 15140-122)
Human EGF (BD Biosciences cat# 354052)
Microsette Six Compartment Biopsy Cassettes (Fisher, 15182705C)
Tools
Lieca VT 1200S Vibrating Microtome (Leica Biosystems, Germany)
Blades (double edge, PTFE coated) (Ted Pella, 121-6)
Adhesive (Ted Pella, 7085-85-0)
Debonder (Ted Pella, 75-52-5)
Rocker Model 55 Variable Speed (Reliable Scientific)
Sterile forceps
Sterile Curved Spatula
Microplate Reader (Molecular Devices, Optimax)
6-mm Disposable Biopsy Punch (Integra-Miltex 33-36)
Preparation of 2% agarose solution
Add 1 gram of low melting point agarose to 25 mL of sterile dPBS in a sterile 250 ml Erlenmeyer flask. Microwave on low power until agarose has completely dissolved. Stop microwave and swirl liquid every 20 seconds so contents do not boil over. Let cool down slightly in 37-degree water bath and then add 25 mL of Williams Medium E. Leave in 37-degree water bath until ready to use.
Preparation of modified WME
Williams’ Media E containing:
nicotinamide (12 mmol/L)
L-ascorbic acid 2-phosphate (50 mg/mL)
D-(+)-and Glucose (5 mg/mL)
Sodium Bicarbonate (2.5%)
HEPES (20 mmol/L)
Sodium Pyruvate (1 mmol/L)
ITS premix (1%)
Glutamax (1%) or L-glutamine (1%)
Penicillin Streptomycin (0.4%)
Human EGF (20 ng/mL)
Procedure
Preparation for slice experiment
- Prepare 5 mL Eppendorf collection tube with 3 mL of Belzer UW Cold Storage Solution. Keep on ice.
- Prepare wash buffer, WME media with 1% Penicillin Streptomycin.
- Prepare modified WME.
- Transfer 400 µL of WME with 1% Penicillin Streptomycin into each well of a 48 well plates, depending on the number of slices desired, and place in tissue culture incubator.
- repare 2% agarose solution and place in 37-degree water bath.
Collection of human tissue
- Criteria for research collection.
- Resection of primary or secondary liver tumors
- Tumor is greater than 2 cm in diameter
- Margins and pathologic assessment will not be compromised from the tissue collection.
- The specimen is placed on the sterile field immediately upon completion of the resection. In the presence of a pathology representative, a wedge of tumor tissue of ~1 cm thick is removed from the specimen using sterile instruments. This is done without sacrificing or violating any potential tissue or margins that are needed for clinical evaluation.
- The tumor tissue wedge is cored using the 6-mm biopsy punch. Using non-tooth pick-ups, cores are placed directly in 3 mL of ice-cold Belzer UW Cold Storage Solution in 5 mL Eppendorf tubes and placed on ice. To maximize tissue viability, cores are taken from the periphery of the tumor to avoid areas of central necrosis.
- Tissue is transported on ice immediately to the lab for processing. These steps are usually completed within 10 minutes of surgical resection.
Preparation of tissue
- Place buffer tray in holder and cover. Add ice around buffer tray.
- Tissue cores are removed from UW solution with sterile forceps and placed in a sterile petri dish.
- Excess liquid is dabbed off on sterile gauze.
- Tissue cores can be directly glued to the specimen disc with adhesive for slicing.
- Longer cores can be cut in half with sterile scalpel.
- Or cores can be embedded in 2% agarose solution for slicing.
- Embedding in agarose is important for softer tissue cores.
- Position core in the center of one well of a sterile 24 well plate.
- Pipet 37 °C, 2% agarose solution over top of core(s) in each well until tissue is entirely covered, place cover on plate, and place on ice.
- Agarose will solidify in less than 5 min.
- Once the agarose has solidified use a sterile scalpel or spatula to cut along the edge of the agarose in the well. With a scooping motion pop the embedded tissue course out of the well and onto a petri dish.
- Using a drop of superglue, glue the face of the embedded core that was on the bottom of the well directly to the specimen disc. Ensure that both the tissue and agarose are in contact with the adhesive. The top of the agarose will have a concave shape.
- Up to 4 cores in agarose or 6 not it agarose can be glued to the specimen disc at one time for efficient slicing.
- The specimen disc is then placed in the buffer tray and the buffer tray is filled with ice cold Belzer UW solution.
Preparation of tumor slices
- Place the ice tray onto the vibratome and secure.
- Rotate blade to position 2.
- Raise the stage up using the control pad until the blade touches the liquid in the buffer tray.
- Set cutting window using the control pad.
- Once the window is set, using the run feature step down 250 microns at a time at a 1.5 mm/s until the blade reaches the tallest tissue core.
- Adjust settings on the vibratome depending on the consistency and integrity of the tissue (amplitude 2–3 mm, speed 0.5–1.5 mm/s).
- Once uniform 250 µm thick slices are obtained, gently scoop out of the bath using a sterile spatula or forceps.
- Place each slice in one well of a 48 well dish containing prewarmed modified WME media.
- Each core and slice were assigned a numerical value so that the sequence and orientation of slices were tracked and documented.
- Once a 48 well plate was filled with slices it was placed at 37 degrees on a rocker (20 rocks/minute) for 1–4 hours for washing.
Determination of viability, MTS assay
- Thaw 5 mL aliquots of CellTiter 96 Aqueous One Solution in the 37-degree water bath.
- Place 400 µL of modified WME into each well of a 48 well dish.
- Transfer slices to individual wells. Keeping track of slice identification number. Keep 3 wells without slices for blanks.
- Add 80 µL of CellTiter 96 Aqueous One Solution to each well of the plate.
- Incubate plate on rocker for 3 hours. (Note: media and slice will change purple).
- Transfer 200 µL of each blank and each slice well to a 96 well assay plate.
- Read plate at 490 nm, blanks are averaged and subtracted from the values of the slices.
- At this point slices can be fixed or transferred back to initial Millicell insert for further culturing.
Preparation of tissue for histology
- Tissue slices are fixed in 400 µL of 10% formalin in a 48 well dish at 4 °C for 24 hours.
- Tissue slices were placed in one compartment each of a six compartment microsette biopsy cassette. Cassettes were labeled with identification code of each slice in each compartment. Cassettes were placed in 70% ethanol.
- Tissue was embedded in paraffin in the orientation placed in the cassette and 4 µm sections were cut and mounted on slides.
Acknowledgments
Funding: Supported by DoD grant W81XWH-16-1-0149 (RSY, QT, VGP).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors have accounted for all aspects of the work including full data access, integrity of the data and the accuracy of the data analysis, and in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Patients undergoing liver resection for primary or metastatic tumors were consented for tissue donation under IRB-approved protocols (University of Washington IRB #1852 and #1666).
References
- Marquart J, Chen EY, Prasad V. Estimation of the Percentage of US Patients With Cancer Who Benefit From Genome-Driven Oncology. JAMA Oncol 2018;4:1093-8. [Crossref] [PubMed]
- Arnedos M, Vicier C, Loi S, et al. Precision medicine for metastatic breast cancer--limitations and solutions. Nat Rev Clin Oncol 2015;12:693-704. [Crossref] [PubMed]
- Vlachostergios PJ, Faltas BM. Treatment resistance in urothelial carcinoma: an evolutionary perspective. Nat Rev Clin Oncol 2018;15:495-509. [Crossref] [PubMed]
- Morash M, Mitchell H, Beltran H, et al. The Role of Next-Generation Sequencing in Precision Medicine: A Review of Outcomes in Oncology. J Pers Med 2018;8. [Crossref] [PubMed]
- Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373:726-36. [Crossref] [PubMed]
- Xu H, Lyu X, Yi M, et al. Organoid technology and applications in cancer research. J Hematol Oncol 2018;11:116. [Crossref] [PubMed]
- Sia D, Moeini A, Labgaa I, et al. The future of patient-derived tumor xenografts in cancer treatment. Pharmacogenomics 2015;16:1671-83. [Crossref] [PubMed]
- Manuelidis L, Manuelidis EE. Studies with electron microscopic autoradiography of thyroxine 125 I in organotypic cultures of the CNS. I. Fixation of thyroxine 125 I. Yale J Biol Med 1972;45:487-500. [PubMed]
- Majumder B, Baraneedharan U, Thiyagarajan S, et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat Commun 2015;6:6169. [Crossref] [PubMed]
- Merz F, Gaunitz F, Dehghani F, et al. Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro Oncol 2013;15:670-81. [Crossref] [PubMed]
- Naipal KA, Verkaik NS, Sanchez H, et al. Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer 2016;16:78. [Crossref] [PubMed]
- Sönnichsen R, Hennig L, Blaschke V, et al. Individual Susceptibility Analysis Using Patient-derived Slice Cultures of Colorectal Carcinoma. Clin Colorectal Cancer 2018;17:e189-99. [Crossref] [PubMed]
- Jiang X, Seo YD, Chang JH, et al. Long-lived pancreatic ductal adenocarcinoma slice cultures enable precise study of the immune microenvironment. Oncoimmunology 2017;6:e1333210. [Crossref] [PubMed]
- Kenerson HL, Aicher LD, True LD, et al. Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res 2002;62:5645-50. [PubMed]
- Kirby TO, Rivera A, Rein D, et al. A novel ex vivo model system for evaluation of conditionally replicative adenoviruses therapeutic efficacy and toxicity. Clin Cancer Res 2004;10:8697-703. [Crossref] [PubMed]
- Webb A, Scott-Mackie P, Cunningham D, et al. The prognostic value of CEA, beta HCG, AFP, CA125, CA19-9 and C-erb B-2, beta HCG immunohistochemistry in advanced colorectal cancer. Ann Oncol 1995;6:581-7. [Crossref] [PubMed]
- Seo YD, Jiang X, Sullivan KM, et al. Mobilization of CD8+ T cells via CXCR4 blockade facilitates PD-1 checkpoint therapy in human pancreatic cancer. Clin Cancer Res 2019. [Crossref] [PubMed]



