Mechanistic study on the inhibition of Staphylococcus epidermidis biofilm by agrC-specific binding polypeptide
Introduction
Staphylococcus epidermidis had emerged as a major opportunistic pathogen in hospitals and is particularly associated with hospital-acquired infections from implanted biomedical materials (1) Implantation of biomedical materials in immunocompromised individuals may facilitate the entry of S. epidermidis into their bodies, gradually leading to bacterial biofilm formation and the subsequent increase in bacterial resistance to exogenous influencing factors (2,3). Biofilm formation occurs in four stages, namely, attachment, aggregation, maturation, and dispersion. Targeting any of these developmental stages may retard the process of biofilm formation.
The accessory gene regulator (agr) quorum sensing system is the most important master regulatory system in S. epidermidis, as it not only participates in the regulation of the initial attachment, aggregation, biofilm maturation, and dispersion stages but also simultaneously regulates the synthesis of virulence factors. In the agr quorum-sensing system, agrC plays a vital role by encoding the sensor AgrC protein (4-6).
In this study, we extracted the genome of S. epidermidis ATCC 35984 strain and performed PCR to amplify the gene fragment of the accessory gene regulator agrC. The T-A clone was added to create a recombinant agrC cloning vector, and the agrC gene was ligated to the pET28α (+) to obtain agrC prokaryotic expression vector. Phage display was used to identify a binding polypeptide unique to agrc. After that, we studied the possible mechanisms underlying the effect of S. epidermidis agrC-specific binding polypeptide on biofilm formation, including the action on the expression of biofilm formation-related genes (atlE, icaA, fbe, and icaR) and synthesis of a biofilm-related product, polysaccharide intercellular adhesin (PIA). This may supply more information for the development of polypeptide drugs specific for the treatment of S. epidermidis biofilm-related infections.
Methods
Bacterial strains, reagents, and equipment
The S. epidermidis strain ATCC 35984 was bought from the American Type Culture Collection (ATCC). The pET-28a (+) was bought from Novagen Inc. The T-Vector pMD19 (simple) was bought from Takara Inc. The E. coli DH5α Competent Cells and BL21(DE3) Competent Cells were obtained from Beijing Solarbio Science & Technology Co., Ltd. The agrC-specific binding polypeptide N1 and agrC-unrelated peptide N0 were synthesized by Shanghai Biotech BioScience & Technology Co., Ltd. Primer was synthesized by Sangon Biotech [Shanghai] Co., Ltd. Eastep® Super Total RNA extraction kit was obtained from Shanghai Promega Biological Products Co., Ltd and iScriptTM cDNA Synthesis kit, from Bio-Rad Inc. The Talent quantitative polymerase chain reaction (qPCR) PreMix (SYBR Green) was supplied by TIANGEN Biotech (Beijing) Co., Ltd, while the Centriprep YM-10 centrifugal filtration unit was obtained from Merck Millipore Inc. The PureLink™ Genomic DNA Mini Kit, PureLink™ HiPure Plasmid Miniprep Kit, multi-functional microplate reader (1510), EASY-nLC1200, Q-Exactive, and fluorescence quantitative PCR machine (ABI-7500) were all purchased from Thermo Fisher Scientific Inc.
Preparation of agrC-specific binding peptide
PureLink™ Genomic DNA Mini Kit was used to extract the genome of S. epidermidis ATCC 35984 strain. PCR was performed to amplify the gene fragment of agrC (The sequences of the primers are shown in Table 1). The purified agrC gene was ligated into the T-vector pmd-19 (simple), and the recombinant plasmid was transformed into E. coli DH5α competent cells. The ampicillin-LB agar medium was used for screening transformed cells. And recombinant agrC cloning plasmid was extracted with PureLink™ HiPure Plasmid Miniprep Kit. The recombinant agrC cloning plasmid was digested by Nhe I and Xho I, and directly ligated to Nhe I and Xho I digested pET-28α (+). The recombinant plasmid was transformed into BL21 (DE3). After purified by the Kanamycin-LB broth medium, we obtained the agrC prokaryotic expression vector. Phage display was used to find agrC-specific binding polypeptide N0 and N1.

Full table
Preparation of the bacterial suspension
Frozen S. epidermidis standard strain ATCC 35984 was dissolved in sterile tryptic soy broth (TSB) culture medium before inoculation on Mueller-Hinton (MH) agar plates. The plates were incubated in a 37 °C thermostatic incubator for 24 h. A single colony was inoculated onto a centrifuge tube holding sterile TSB culture medium and cultured at 37 °C and 150 rpm for 16–18 h. The bacterial cells could grow to the logarithmic growth phase, and the concentration of cells in the suspension was adjusted to 1×107 CFU/mL.
Effects of different concentrations of the agrC-specific binding polypeptide on S. epidermidis biofilm
N1 and N0 polypeptide powders were dissolved in sterilized deionized water to obtain N1 and N0 polypeptide solutions, respectively. Polypeptides N1 and N0 have been used to treat bacterial suspension at final concentrations of 0, 100, 200, 400, 800, and 1,600 µg/mL. Bacterial suspensions treated with different concentrations of polypeptides were added to a 96-well cell culture plate and cultured for 12 h in a 37 °C thermostatic incubator. There was crystal-violet staining, and the absorbance at 570 nm wavelength (OD570) was measured to determine S. epidermidis biofilm formation ability in the presence of different concentrations of polypeptides.
Effects of agrC-specific binding polypeptide treatment for different time points on S. epidermidis biofilm formation ability
Polypeptide solutions N1 and N0 were added to the bacterial suspensions at a final concentration of 800 µg/mL. Bacterial suspensions treated with N1 and N0 polypeptide solutions were appointed as N1 and N0 groups, respectively. An equal volume of sterile deionized water without polypeptide was added to the control group. Bacterial suspensions from the various groups were added to 24-well cell culture plates, which were then incubated at 37 °C. One of the 24-well cell culture plates was removed from each group at 6, 12, 18, 24, and 30 h of culture and treated with 400 mL of TSB culture medium and 80 µL XTT solution (added to each well). The plates were incubated in the dark at 37 °C for 2 h. Following incubation, each well had a solution removed, and its absorbance at 450 nm wavelength was measured for the staining removal solution in each well
Quantitation of biofilm-related gene expression
One 24-well cell culture plate was removed from all groups at each time point, and the biofilms were scraped and transferred to 1.5 mL centrifuge tubes. The Eastep®super total RNA extraction kit was used to extract total RNA from S. epidermidis for RNA quantitation. Reverse transcription was conducted with the extracted RNA samples on a PCR machine. Primers were designed for atlE, icaA, fbe, icaR, and 16 sRNA, which was used as the reference gene (The sequences of the primers are shown in Table 2). The primers and cDNA template synthesized from the reverse transcription reaction were used for the real-time (RT)-qPCR reaction. The 2−∆∆Ct method was used for the comparison of the relative levels of mRNAs.

Full table
Evaluation of PIA synthesis
In the sulfuric acid-phenol method, polysaccharides are first hydrolyzed to monosaccharides under the action of sulfuric acid before undergoing rapid dehydration to form uronic acid. Uronic acid then reacts with phenol to form an orange-yellow compound, which was then colorimetrically quantified.
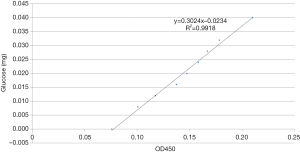
One 24-well cell culture plate was removed from each group at different time points. The biofilms were scraped and transferred to 1.5 mL centrifuge tubes. After centrifugation, the supernatant was discarded, and the cells in deionized water were resuspended. The bacterial cells were sonicated (sonication for 8 s, followed by 1 s of rest for a total of 5 min). In total, 15 µL of 37% formaldehyde was added to each tube and evenly mixed. We incubated the tubes at 4 °C for 2 h and treated with 200 µL sodium hydroxide (NaOH; 1 mol/L) at 4 °C for 3 h. The tubes were centrifuged, and the supernatant discarded. The pellet was resuspended in 1 mL of deionized water and treated with 0.5 mL of 5% phenol and 2.5 mL of concentrated sulfuric acid. The reactants were evenly mixed and allowed to cool. The absorbance at 490 nm (OD490) was measured after incubation of the samples at 25 °C for 20 min. We used 0, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, and 1.0 mL glucose solutions (40 mg/mL) to plot a standard curve. Deionized water was added to make up the volume to 1.0 mL, and the method was used to measure OD490. The OD values were used as the y-axis coordinates, and the corresponding glucose levels were used as x-axis coordinates to plot a standard curve.
Statistical analysis
The SPSS 24.0 statistical software was used for analysis. Analysis of variance (ANOVA) was used for intragroup and intergroup comparison, and the t-test was used for pairwise comparison. A value of P<0.05 was considered statistically significant.
Results
Obtain of agrC specific binding peptide
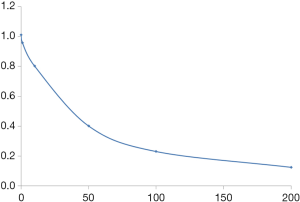
By analyzing the phage, we obtained two peptide sequences (Table 3), but only N1 showed a strong inhibition effect (Figure 1).

Full table
Effects of different concentrations of the agrC-specific binding polypeptide on S. epidermidis biofilms
At polypeptide concentrations lower than 400 µg/mL, no difference between N1 and N0 groups was found in the OD570 values (P>0.05). Treatment with polypeptide at 800 and 1,600 µg/mL concentrations resulted in a significant decrease in the OD570 value for the N1 group as compared with that for the N0 group (P<0.05). Among N1-treated groups, the group exposed to 800 µg/mL of N1 had significantly lower OD570 value than the groups treated with other concentrations (P<0.05); the differences in OD570 values between groups treated with other concentrations were not statistically significant (P>0.05). The intragroup comparison after N0 treatment revealed no statistically significant difference in OD570 values regardless of the concentrations (P>0.05) (Figure 2).
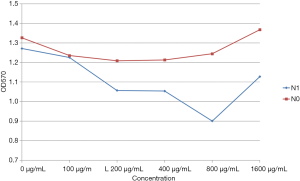
Effects of different concentrations of peptides on biofilm formation ability of Staphylococcus epidermidis ATCC35984.
Effects of agrC-specific binding polypeptide treatment for different time points on S. epidermidis biofilm formation ability
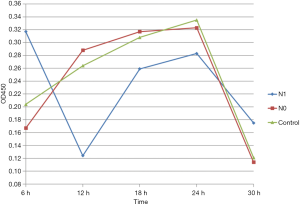
At any time, point, the difference between the N0 group and the control group was not statistically significant (P>0.05). After 6 h of culture, the OD450 value was higher for the N1 group than that for N0 and control groups (P<0.05). However, the OD450 value for the N1 group was lower than that for N0 and control groups after 12 h of culture (P<0.05). No significant differences in OD450 values were noted between distinct groups at other time points (P>0.05) (Figure 3).
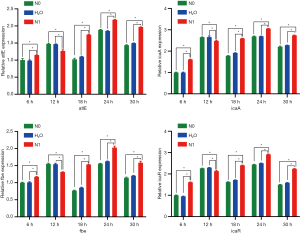
Biofilm-related gene expression
No differences were noted in the expression levels of various genes between N0 and control groups at various time points (P>0.05). After 12 h of culture, the expression levels of atlE, icaA, fbe, and icaR genes were downregulated in the N1 group as compared to those in N0 and control groups (P<0.05). At other time points, the expression of these four genes was higher in the N1 group than in N0 and control groups (P<0.05) (Figure 4).
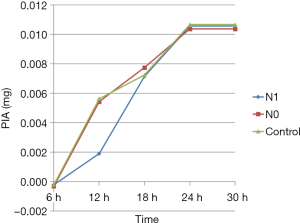
PIA synthesis
The regression formula for the glucose standard curve was y=0.3024x−0.0234 and R2=0.9918 (Figure 5).
The PIA levels in various groups at different time points were calculated from the standard curve. The PIA content was significantly lower in the N1 group than in N0 and control groups (P<0.05) only at 12 h; no statistically significant differences were observed between different groups at other time points (P>0.05). Also, the PIA content in various groups increased with time from 6 to 24 h of culture. However, no significant differences were reported in the PIA content between various groups at 24 and 30 h (Figure 6).
Discussion
In clinical practice, bacterial infections have become normal and the development of biofilms during bacterial infections increases the resistance of the infecting bacteria to external interference, ultimately leading to refractory biofilm-related infections (7). S. epidermidis causes 20–30 per cent of orthopedic device-related infections, and the incidence of late-developing infections may even rise to 50 per cent (8).
S. epidermidis is equipped with two quorum-sensing systems, namely the agr quorum-sensing system and the luxS quorum-sensing system. Quorum sensing systems enable communication between individual bacterial cells through close connections and promote the formation of bacterial biofilms as well as the expression of virulence factors (4,5). The agr quorum-sensing system plays a vital role in S. epidermidis, as it regulates attachment and affects biofilm formation. This system can also regulate bacterial toxin expression and biofilm transfer. The agr system includes two units (RNAII and RNAIII), and their P2 and P3 promoters drive their transcription. RNAII is composed of four genes (agrA, agrB, agrC, and agrD), wherein agrD is transcribed and synthesized into the precursor of autoinducing peptides (AIPs), and agrB encodes for a transmembrane endopeptidase responsible for the processing of AIP precursors to form AIPs. The processed AIPs then undergo cleavage mediated by the type I signal peptidase SpsB. The agrC and agrA genes encode a sensor AgrC and a regulator AgrA in a dual-component signal transduction system. As the level of AIP reaches its threshold value, it activates the phosphorylation or dephosphorylation of AgrC and AgrA, and consequently induces the expression of the promoters P2 and P3, ultimately leading to the transcription of RNAII and RNAIII (4,6).
Many biofilm synthesis genes take part in the formation of S. epidermidis biofilms, including atlE, fbe, and icaA. The multiple secretory signals emitted during bacterial biofilm formation could also aid bacteria in their physiological responses toward a specific environment.
The at1E gene plays a vital role during the initial phase of biofilm formation, as it is intimately associated with bacterial attachment. Autolysin (At1E) encoded by at1E is a 115 kDa protein that serves as a peptidoglycan hydrolase. This enzyme plays a crucial role in the degradation of bacterial cell walls. At the same time, it increases the surface hydrophobicity of the bacterial cell and increases its attachment to polyethylene materials (9). fbe is an important regulatory gene involved in the first phase of biofilm formation. The fibrinogen-binding protein (Fbe) encoded by fbe gene mediates the attachment of S. epidermidis to fibrinogen through specific “dock and lock” mechanisms, resulting in the formation of stable protein-ligand complexes (10,11). A study found that the expression of the fbe gene could promote the attachment of S. epidermidis on fibrinogen-coated biomaterials (12).
The aggregated S. Several genes regulate epidermidis, including the ica locus, which forms five genes, namely icaA, icaB, icaC, icaD, and icaR. The entire length of the icaABCD gene is 3.5 kb and jointly encodes PIA, a major part of the extracellular matrix that mediates cell-cell adhesion in bacteria. The genes icaA and icaD take part in the synthesis of N-acetylglucosaminyltransferase and adds activated N-acetylglucosaminyltransferase to the continuously increasing PIA chain. The product of icaC transports PIA out of the cell, while the product of icaB is responsible for the deacetylation of N-acetylglucosamine in PIA to facilitate the localization, adhesion, and other biological activities of PIA on biological or non-biological surfaces (1,11,13). The icaR gene is found upstream of icaABCD and binds to a specific DNA sequence upstream of the neighboring icaA gene to elicit its effects. Related studies have reported that icaR is a repressor, and its product may decrease PIA yield (14,15).
agrC is a key target molecule in the agr quorum-sensing system. In the present study, we studied the effects of these peptides on biofilm formation-related genes (atlE, icaA, fbe, and icaR) and the biofilm formation-related product PIA to evaluate the possible mechanism underlying the repressive effects on biofilm formation.
In S. epidermidis, the agr gene is 3.5 kb long and forms two independent transcription units (RNAII and RNAIII). RNII consists of 4 genes; namely, agrA, agrB, agrC, and agrD, wherein agrD and agrB interact to form the signaling molecule AIP. AIP then participates in the agrC quorum-sensing system in a concentration-dependent manner; after reaching the threshold concentration, the sensor agrC gets activated and promotes agrA phosphorylation or dephosphorylation, leading to the activation of P2 and P3 promoters and the subsequent transcription of RNAII and RNAIII (3,5,16). Therefore, we deduce that the S. epidermidis agrC-specific binding polypeptide is like AIPs and may specifically bind to agrC in a concentration-dependent manner. We evaluated the ability of bacteria to form biofilms in 12 h after treatment with different concentrations of agrC-specific binding polypeptide. As a result, we found that low concentrations of (400 µg/mL and below) agrC-specific binding polypeptide had no effects on the ability of S. epidermidis to form biofilms. Any further increase in the concentration of agrC-specific binding polypeptide led to the inhibition of biofilm formation ability. At 800 µg/mL concentration, agrC-specific binding polypeptide showed maximum inhibition of S. epidermidis biofilm formation and any further increase in peptide concentration did not affect biofilm formation. This observation shows that the inhibition of S. epidermidis biofilm formation by S. epidermidis agrC-specific binding polypeptide shows a concentration-response relationship and that the final concentration of 800 µg/mL was the optimum dose required to inhibit biofilm formation by S. epidermidis.
The OD450 values were measured using the XTT assay and were positively correlated with the live bacterial count. Therefore, this method can indirectly reflect the viability of bacterial cells at different time points after treatment with the agrC-specific binding polypeptide. After 6 h of cultivation, the viability of bacterial cells was higher in the N1 group than in N0 and control groups. However, after 12 h, the bacterial viability significantly decreased in the N1 group, while the bacteria in N0 and control groups continued to grow. The viability count remained stable for all groups after 18 and 24 h of culture but decreased at 30 h in all groups owing to nutrient depletion. The agrC-specific binding polypeptide showed transient inhibitory effects on S. epidermidis viability only after 12 h of culture and could even promote bacterial attachment at the first stages of growth.
In the present study, we evaluated the expression of the genes that are associated with early attachment in S. epidermidis during biofilm formation, including atlE, icaA, and fbe. The results of RT-qPCR showed that the expression of atlE, icaA, fbe, and icaR was downregulated in the N1 group as compared with that in the N0 group after 12 h of culture. However, the expression levels of these four genes were higher in the N1 group than in the two other groups at other time points. Consistent with the results of XTT assay, we believe that agrC-specific binding polypeptide may upregulate the expression of attachment-related genes after 6 h of culture, thereby promoting the early attachment of S. epidermidis. At 12 h of culture, agrC-specific binding polypeptide transiently downregulated the expression of attachment-related genes and thereby repressed S. epidermidis cell attachment and viability. At 18 h of culture, the upregulation in the expression of attachment-related genes (atlE, icaA, and fbe) could not continue to promote the formation of biofilm, as the process of early attachment was complete and resulted in the promotion of biofilm formation. Therefore, the bacterial viability did not continue to increase even after the upregulation in the expression of attachment-related genes in the N1 group after 18 h of culture.
Among the tested genes, icaR expression upregulation could reduce PIA synthesis, and so inhibit biofilm formation. PIA is a vital part of the extracellular matrix and an important substance for biofilm formation. PIA is coded by icaABCD and icaR. However, RT-qPCR results showed that the expression levels of icaR and icaA were similar, and the sulfuric acid-phenol assay showed that PIA synthesis only decreased at 12 h of culture, consistent with the simultaneous downregulation in icaA expression level. Thus, icaR upregulation caused by agrC-specific binding polypeptide did not produce significant inhibitory effects, while the downregulation of its expression was unable to increase PIA synthesis. Therefore, we suggest that agrC-specific binding polypeptide may not affect the biofilm formation ability of S. epidermidis by regulating icaR expression alone but through the upregulation or downregulation of the expression of ica genes. However, changes in icaR expression may not antagonize the positive or inhibitory effects of icaABCD.
In an earlier study, we successfully synthesized a polypeptide that could specifically bind to agrC. In the present study, we demonstrate that agrC-specific binding polypeptide downregulated the expression of atlE, icaA, fbe, and icaR genes and inhibited PIA formation after cultivation with S. epidermidis for 12 h, indicative of its transient inhibitory effects on the bacterial biofilm formation ability. Its effective concentration was 800 µg/mL. The widespread misuse of antibiotics demands studies on new antimicrobial treatments to delay the development of antibiotic resistance in bacteria. Although our study shows that agrC-specific binding polypeptide could transiently inhibit biofilm formation, this effect may be insufficient to extend its application for the treatment of S. epidermidis infections. Therefore, further studies are called for to enhance and extend the duration of this effect. Also, we only selected some attachment-related genes for expression testing and did not completely clarify other possible antibacterial mechanisms of actions of the agrC-specific binding polypeptide. A complete gene expression tests are still needed to understand the possible mechanisms.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (81860325 and 81460278); National Natural Science Foundation of China (81960335); Natural Science Foundation of Yunnan Province (2017FA039); Natural Science Foundation of Yunnan Province (2017FE468-214); Medical Experts Training Project of Yunnan Province (D-201641 and L-2017006); National key R & D plan of China (2017FYC0907902).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Otto M. Staphylococcus epidermidis pathogenesis. Methods Mol Biol 2014;1106:17-31. [Crossref] [PubMed]
- Lianhua Y, Yunchao H, Guangqiang Z, et al. The effect of iatrogenic Staphylococcus epidermidis intercellar adhesion operon on the formation of bacterial biofilm on polyvinyl chloride surfaces. Surg Infect (Larchmt) 2014;15:768-73. [Crossref] [PubMed]
- Wolska KI, Grudniak AM, Rudnicka Z, et al. Genetic control of bacterial biofilms. J Appl Genet 2016;57:225-38. [Crossref] [PubMed]
- Kavanaugh JS, Horswill AR. Impact of Environmental Cues on Staphylococcal Quorum Sensing and Biofilm Development. J Biol Chem 2016;291:12556-64. [Crossref] [PubMed]
- Laverty G, Gorman SP, Gilmore BF. Biomolecular mechanisms of staphylococcal biofilm formation. Future Microbiol 2013;8:509-24. [Crossref] [PubMed]
- Le KY, Otto M. Quorum-sensing regulation in staphylococci-an overview. Front Microbiol 2015;6:1174. [Crossref] [PubMed]
- Yong YY, Dykes GA, Choo WS. Biofilm formation by staphylococci in health-related environments and recent reports on their control using natural compounds. Crit Rev Microbiol 2019;45:201-22. [Crossref] [PubMed]
- Sabaté Brescó M, Harris LG, Thompson K, et al. Pathogenic Mechanisms and Host Interactions in Staphylococcus epidermidis Device-Related Infection. Front Microbiol 2017;8:1401. [Crossref] [PubMed]
- Büttner H, Mack D, Rohde H. Structural basis of Staphylococcus epidermidis biofilm formation: mechanisms and molecular interactions. Front Cell Infect Microbiol 2015;5:14. [PubMed]
- Vuong C, Gerke C, Somerville GA, et al. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis 2003;188:706-18. [Crossref] [PubMed]
- Paharik AE, Horswill AR. The Staphylococcal Biofilm: Adhesins, Regulation, and Host Response. Microbiol Spectr 2016. [Crossref] [PubMed]
- Vanzieleghem T, Herman-Bausier P, Dufrene YF, et al. Staphylococcus epidermidis Affinity for Fibrinogen-Coated Surfaces Correlates with the Abundance of the SdrG Adhesin on the Cell Surface. Langmuir 2015;31:4713-21. [Crossref] [PubMed]
- Arciola CR, Campoccia D, Ravaioli S, et al. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front Cell Infect Microbiol 2015;5:7. [Crossref] [PubMed]
- Hoang TM, Zhou C, Lindgren JK, et al. Transcriptional Regulation of icaADBC by both IcaR and TcaR in Staphylococcus epidermidis. J Bacteriol 2019. [Crossref] [PubMed]
- Yu D, Zhao L, Xue T, Sun B. Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC Microbiol 2012;12:288. [Crossref] [PubMed]
- Yang T, Tal-Gan Y, Paharik AE, et al. Structure-Function Analyses of a Staphylococcus epidermidis Autoinducing Peptide Reveals Motifs Critical for AgrC-type Receptor Modulation. ACS Chem Biol 2016;11:1982-91. [Crossref] [PubMed]