
CPS1 expression and its prognostic significance in lung adenocarcinoma
Introduction
Malignant lung cancer tumors are the most common cause of cancer-related deaths worldwide. Every year, countless patients worldwide die from this disease (1). Among them, the incidence of lung adenocarcinoma (LADC), the most common histological subtype of lung cancer, is increasing year by year (2,3). Due to the delay in diagnosis, however, the treatment effect for LADC is strongly unsatisfactory (4,5). Despite the emerging advances in diagnostic and therapeutic techniques, it remains a serious global public health concern that is characterized by a lack of effectual progress in advanced diagnosis and treatment (6). Therefore, it is necessary to identify novel molecular markers to improve the early diagnosis and treatment of LADC.
The enzyme carbamoyl phosphate synthetase 1 (CPS1) forms carbamoyl phosphate from bicarbonate, ammonia, and adenosine triphosphate (ATP) and is activated allosterically by N-acetylglutamate. The neonatal presentation of bi-allelic mutations of CPS1 results in hyperammonemia with reduced citrulline (7). Moreover, CPS1 is the rate-limiting enzyme in the first step of the urea cycle and an indispensable enzyme in human liver metabolism (8). Therefore, CPS1 is closely related to liver disease, including evolution of chronic HCV infection and hepatic fibrosis (9). In addition, emerging studies have shown that the metabolic gene CPS1 exhibits significantly reduced expression in poorly differentiated hepatocellular carcinoma cell lines (10). Another report also indicates that the overexpression of CPS1 is correlated with both of the adverse therapeutic responses in colorectal cancer patients receiving neoadjuvant concurrent chemoradiotherapy (CCRT) (11). At the same time, Pham-Danis et al. recently found that downregulation of CPS1 with EGFR inhibitors could further reduce the proliferation of EGFR-mutant non-small cell lung cancer (NSCLC) cells and prevent cell cycle progression (12). However, the specific role of CPS1 in LADC and its detailed mechanisms necessitate further study.
The purpose of our study was to investigate the roles of CPS1 in human LADC and its relationship with clinical treatment and prognosis, so as to provide a new option for further effective treatment of LADC patients.
Methods
Data collection and reanalysis using different bioinformatics methods
We summarize several bioinformatics databases used to evaluate the CPS1 expression levels in LADC tissues and cell lines in Table S1.

Full table
The differential biomarker screening and clinicopathologic based analysis could be carried out using the Oncomine database containing 65 gene expression datasets from most major types of cancer, along with their normal tissues and various cancer subtypes (13). We performed an exploration of CPS1 expression between LADC and normal tissues. In addition, two databases, Gene Expression Profiling Interactive Analysis (GEPIA) (14) and UALCAN (15), were used to validate the results. Through these public bioinformatics webtools, we could clearly understand the expression profiles of CPS1 in human LADC tissues and cell lines.
Wanderer is an intuitive webtool allowing real-time access and visualization of gene expression and DNA methylation profiles from The Cancer Genome Atlas (TCGA) (16). This allows us to screen for possible methylation sites in the full sequence of CPS1 DNA and to analyze the associations between clinical features of LADC patients, CPS1 expression and its methylation values.
Kaplan-Meier survival analysis was used to assess the impact of genes on patient survival (17). We used Kaplan-Meier Plotter to describe the relationships between CPS1 expression levels and first-progression (FS) and postprogression survival time (PPS) (18). Otherwise, the determination of associations between CPS1 expression and disease-free survival (DFS) and overall survival (OS) was achieved through the GEPIA and UALCAN databases, respectively.
The Gene Expression Omnibus (GEO) database is an international public repository that archives and freely distributes high-throughput gene expression and other functional genomics datasets (19). We obtained two therapeutically relevant transcriptome microarrays, GSE2189 (20) and GSE54712 (21), from the GEO database. Subsequently, the effect of CPS1 expression on LADC chemotherapy was analyzed.
cBioPortal, a web-based resource for analyzing multidimensional cancer genomics data (22), was used to screen for genes coexpressed with CPS1 in LADC tissues. Furthermore, we used a STRING database (23) and Cytoscape tool (24) to construct a protein-protein interaction network (PPI) of these coexpressed genes. We then performed Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the CPS1 coexpressed genes in LADC samples using the web-based GEne SeT AnaLysis toolkit (WebGestalt) (25) and Pathview algorithm (26), respectively.
MethHC is a database comprising a systematic integration of DNA methylation data and mRNA/microRNA expression profiles in human cancers (27). We use it to evaluate the correlation analysis between CPS1 expression and its methylation values. For the correlation between disease prognosis and CPS1 methylation values, the MethSurv tool was used to examine the effect of CPS1 methylation on patient survival time (28).
Statistical analyses
We used Student’s t-test and statistical package SPSS (SPSS) 12.0 (IBM Analytics) to analyze genes differentially expressed between cancerous and noncancerous tissues. The relationship between CPS1 expression and clinicopathological features was also analyzed using ANOVA and K independent sample tests, and correlations between genes were assessed using Pearson correlation coefficients. Meanwhile, significance of survival analysis was performed using Kaplan-Meier analysis with the Log-rank test. P<0.05 was considered to be statistically significant.
Results
CPS1 is upregulated in LADC tissues and cell lines
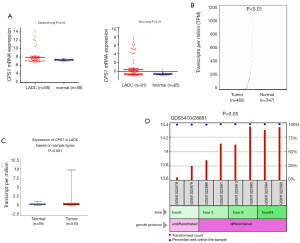
To examine changes in CPS1 expression between LADC and adjacent nontumor tissues, we analyzed their expression profiles in a number of independent bioinformatics databases. Using the Oncomine database, we found that the CPS1 transcription levels in tumor tissues from two individual microarray datasets increased significantly (Figure 1A). To further confirm these results, we analyzed the expression of CPS1 in LADC using GEPIA and UALCAN: we obtained the same change trends as above (Figure 1B,C). Expectedly, we obtained a GSE54712 dataset (21) from the GEO database and found that CPS1 could induce tumor development and metastasis under the synergy of cancer stem cells (CSCs) (Figure 1D). All of the above data indicated that the upregulation of CPS1 expression level promotes the development and progression of LADC.
CPS1 expression is associated with the clinical characteristics of LADC patients
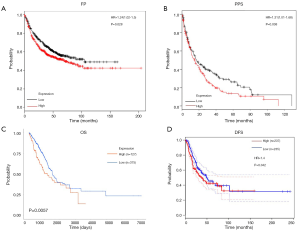
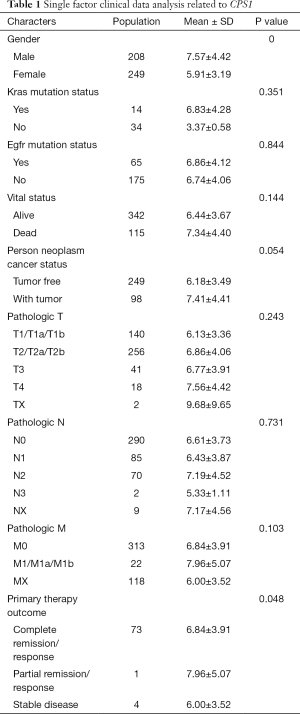
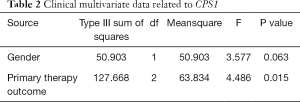
To date, almost no literature has reported a relationship between the expression of CPS1 and the clinical prognosis of human LADC. The effect of CPS1 expression on survival index was assessed using a Kaplan-Meier plotter tool, confirming that upregulation of CPS1 expression was significantly associated with shorter FP and PPS (P=0.028 and 0.038, respectively) (Figure 2A,B). Furthermore, the UALCAN database showed that patients with high CPS1 levels had shortened OS (P=0.0057) (Figure 2C). At the same time, from the GEPIA database, we also found that patients with high levels of CPS1 expression had shorter DFS (P=0.042) (Figure 2D). Using the Wanderer database, we obtained a series of clinical data, summarized as Table 1. Correlations between CPS1 expression with gender (P=0.000) and primary therapy outcome (P=0.048) are shown in Table 1. No correlation was observed between CPS1 expression and other clinical features, such as KRAS mutation status, EGFR mutation status, etc. Multivariate analysis using the COX regression model showed that the primary therapy outcome was independently associated with CPS1 transcription levels in LADC samples (Table 2). In conclusion, CPS1 could serve as a potential biomarker for diagnosis and prognosis of LADC patients.
Full table
Full table
The role of CPS1 in LADC therapies
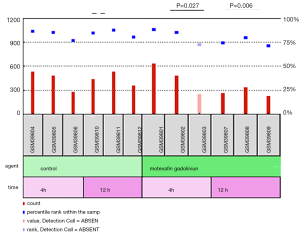
Next, we screened the chemotherapy-related microarray datasets from the GEO database to further determine the roles of CPS1 in the treatment of LADC patients. From the GSE2189 dataset (20), we found that the expression level of CPS1 was significantly decreased at 4 and 12 hours after treatment with the chemotherapy drug motexafin in the A549 human lung cancer cell line (P<0.05) (Figure 3). These findings indicated that changes in CPS1 expression levels might be related to cancer treatment response.
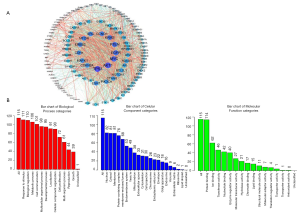
Functional enrichment analysis of CPS1 coexpressed genes
We downloaded the information of genes coexpressed with CPS1 using the cBioPortal database and obtained 174 coexpressing differential genes (co-DEGs) (http://fp.amegroups.cn/cms/d7e742548b77ff1fe1b3d13978dec587/atm.2020.02.146-1.doc). The PPI networks of these 174 co-DEGs were then constructed by STRING and Cytoscape (Figure 4A). To further understand the biological functions of these co-DEGs, we performed GO annotation and KEGG pathway analysis. The GO annotation identifies the major biological processes (response to stimulus and biological regulation), cellular components (nucleus) and molecular functions (protein binding) of CPS1 biology (Figure 4B). The Pathview database was used to analyze the KEGG pathway: as shown in Table S2, the corresponding pathways (osteoclast differentiation, regulation of actin cytoskeleton, apoptosis) were obtained.

Full table
Relationship between CPS1 methylation and clinical features of patients with LADC
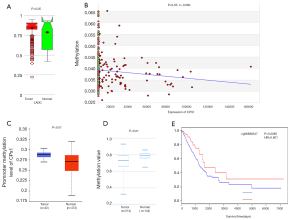
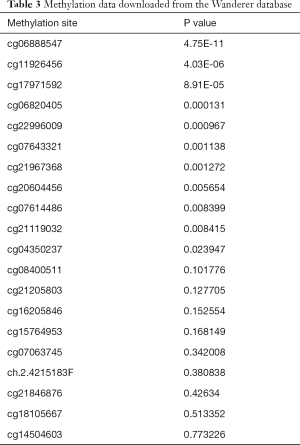
It is well known that there is a negative correlation between DNA methylation and gene expression (29-31). From the MethHC database, we observed that the global CPS1 methylation level in the LADC samples was lower than that in normal samples (P<0.05) (Figure 5A) and was negatively correlated with expression (P<0.05, r=−0.080) (Figure 5B), supporting the high expression of CPS1 in LADC samples. Next, we used the UALCAN and DiseaseMeth databases to further validate the results of CPS1 methylation level. The results showed that CPS1 presented low methylation levels in human LADC patients (Figure 5C,D). Subsequently, we screened the methylation site cg06888547 from the Wanderer database as the most statistically significant candidate site (Table 3). Using the MethSurv network tool analysis, it was found that higher methylation values of cg06888547 in LADC patients were associated with longer OS (P=0.0085) (Figure 5E). These data demonstrated that DNA hypomethylation, a major epigenetic modification, might lead to CPS1 overexpression at the transcriptional level, thus plays important roles in carcinogenesis and progression of LADC patients.
Full table
Discussion
Our research first has found that CPS1 can be used as a proto-oncogene of LADC and can be used as a potential biomarker through comprehensive mining of public databases. We also analyzed the differential coexpression of CPS1 and found a possible signaling pathway for CPS1 to determine its biological significance in cancer development. The Oncomine, UALCAN and GEPIA datasets indicate that CPS1 is highly expressed in LADC. Kaplan-Meier survival analysis also showed that patients with elevated CPS1 levels had shortened OS and PPS. In addition, the methylation level of CPS1 is opposite to that of protein expression, showing a low level.
CPS1 is the first rate-limiting mitochondrial enzyme in the urea cycle. Lack of CPS1 due to mutation can lead to life-threatening hyperammonemia (32,33). A recent study by Çeliktas et al. has shown that knockdown of CPS1 induced accumulation of ammonia and reduction of nucleic acid synthesis pathway, leading to the decreased cancer cell growth. At the same time, they have proven the increased of CPS1 expression was statistically related with the worse OS both at mRNA and protein levels, which is consistent with what we found (32). Therefore, CPS1 may become a potential molecular biomarker of poor prognosis and facilitate the individualized treatment for LADC. In addition, in hepatocellular carcinoma (HCC), CPS1 is downregulated after treatment with aflatoxin B1 (AFB1), an effective hepatocarcinogen, and may serve a differentiation function (34). The researchers discovered the hypermethylation and decreased RNA expressions of CPS1 in most HCC patients. They speculated that CPS1 might serve an inhibitory action in the tumorigenesis and development of HCC cancer cells (35). However, our study indicated that CPS1 might be an oncogenic factor for LADC biology, which may be due to the activated Janus kinases (JAKs)/Signal transducers and activators of transcription (STAT) signaling pathway (32). Generally, JAK-STAT signaling activation has been proved to promote cancer progress through modulating the inflammation, invasion and immunosuppression (36).
Although a large body of literature indicates that CPS1 plays a crucial role in human malignancies, little is known about the detailed roles of CPS1 in human LADC. Our research has fully demonstrated that CPS1 promotes tumors in LADC samples. Moreover, we have demonstrated that patients with high expression of CPS1 have worse OS and PPS. This provides an idea for further exploration of CPS1 as a potential biomarker for LADC. Human LADC tumorigenesis is impacted by epigenetic changes, a situation which contributes to the abnormal changes of specific genes. Moreover, the other promising cause for LADC diagnostics could be the methylation values of DNA sequences in certain biomarkers (37). To date, the most meaningful and common epigenetic modifications in the mammalian genome are DNA methylation events. Oshima et al. found that the methylation of metastasis-suppressive miRNAs could upregulate these miRNA expression levels, leading to the metastasis profiles of cancer cells in lung carcinogenesis (38). DNA hypermethylation-mediated downregulation of LINC00261 plays an important proliferation-inducing role in LADC progression (39). By reanalyzing the datasets from several user-friendly webtools, we found the significant low-methylated values of CPS1 and identified the negative association between its methylation and expression levels, thus confirming that DNA hypomethylation was indeed responsible for the increased CPS1 expression in LADC tissues.
However, our study has several limitations. First of all, our research is mainly based on the analysis of biological databases, lacking effective external experimental validation. Secondly, it is necessary to include more prognostic variables to improve the accuracy of survival analysis. Finally, further validation of CPS1 in multicenter clinical trials and prospective studies is much needed.
Conclusions
Taken together, our results indicated that CPS1 is a candidate tumor proto-oncogene for human LADC. The chemotherapy results of LADC also indicate that CPS1 is a potential indicator of cancer treatment efficacy. In addition, reanalysis based on public databases provides new insights into the screening of potential biomarkers associated with human malignant diseases, especially human lung cancers.
Acknowledgments
Funding: This work was supported by Key Research and Development Program of Hunan Province (No. 2018SK2091), the National Natural Science Foundation of China (No. 81703036, 81803035), the China Postdoctoral Science Foundation (No. 2017M610510), the Natural Science Foundation of Hunan Province (2019JJ50932), the Youth Fund of Xiangya Hospital (No. 2017Q17), and the Fundamental Research Funds for the Central Universities of Central South University (2019zzts800, 2019zzts345).
Footnote
Conflicts of Interest: The authors have no conflict of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yan Y, Xu Z, Qian L, et al. Identification of CAV1 and DCN as potential predictive biomarkers for lung adenocarcinoma. Am J Physiol Lung Cell Mol Physiol 2019;316:L630-43. [Crossref] [PubMed]
- DeSantis CE, Miller KD, Dale W, et al. Cancer statistics for adults aged 85 years and older, 2019. CA Cancer J Clin 2019;69:452-67. [Crossref] [PubMed]
- Wei J, Yan Y, Chen X, et al. The Roles of Plant-Derived Triptolide on Non-Small Cell Lung Cancer. Oncol Res 2019;27:849-58. [Crossref] [PubMed]
- Yan Y, Su W, Zeng S, et al. Effect and Mechanism of Tanshinone I on the Radiosensitivity of Lung Cancer Cells. Mol Pharm 2018;15:4843-53. [Crossref] [PubMed]
- Zhou Y, Hoti N, Ao M, et al. Expression of p16 and p53 in non-small-cell lung cancer: clinicopathological correlation and potential prognostic impact. Biomark Med 2019;13:761-71. [Crossref] [PubMed]
- Zhou M, Wang H, Zeng X, et al. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2019;394:1145-58. [Crossref] [PubMed]
- Khoja S, Nitzahn M, Truong B, et al. A constitutive knockout of murine carbamoyl phosphate synthetase 1 results in death with marked hyperglutaminemia and hyperammonemia. J Inherit Metab Dis 2019;42:1044-53. [Crossref] [PubMed]
- Chen Z, Tang N, Wang X, et al. The activity of the carbamoyl phosphate synthase 1 promoter in human liver-derived cells is dependent on hepatocyte nuclear factor 3-beta. J Cell Mol Med 2017;21:2036-45. [Crossref] [PubMed]
- El-Sheikh RM, Mansy SS, Nessim IG, et al. Carbamoyl phosphate synthetase 1 (CPS1) as a prognostic marker in chronic hepatitis C infection. APMIS 2019;127:93-105. [Crossref] [PubMed]
- Nwosu ZC, Battello N, Rothley M, et al. Correction to: Liver cancer cell lines distinctly mimic the metabolic gene expression pattern of the corresponding human tumours. J Exp Clin Cancer Res 2018;37:267. [Crossref] [PubMed]
- Lee YY, Li CF, Lin CY, et al. Overexpression of CPS1 is an independent negative prognosticator in rectal cancers receiving concurrent chemoradiotherapy. Tumour Biol 2014;35:11097-105. [Crossref] [PubMed]
- Pham-Danis C, Gehrke S, Danis E, et al. Urea Cycle Sustains Cellular Energetics upon EGFR Inhibition in EGFR-Mutant NSCLC. Mol Cancer Res 2019;17:1351-64. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Díez-Villanueva A, Mallona I, Peinado MA. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin 2015;8:22. [Crossref] [PubMed]
- Yan Y, Xu Z, Hu X, et al. SNCA Is a Functionally Low-Expressed Gene in Lung Adenocarcinoma. Genes (Basel) 2018;9. [Crossref] [PubMed]
- Yan P, He Y, Xie K, et al. In silico analyses for potential key genes associated with gastric cancer. PeerJ 2018;6:e6092. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Magda D, Lecane P, Miller RA, et al. Motexafin gadolinium disrupts zinc metabolism in human cancer cell lines. Cancer Res 2005;65:3837-45. [Crossref] [PubMed]
- Lopez-Ayllon BD, Moncho-Amor V, Abarrategi A, et al. Cancer stem cells and cisplatin-resistant cells isolated from non-small-lung cancer cell lines constitute related cell populations. Cancer Med 2014;3:1099-111. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Morris JH, Knudsen GM, Verschueren E, et al. Affinity purification-mass spectrometry and network analysis to understand protein-protein interactions. Nat Protoc 2014;9:2539-54. [Crossref] [PubMed]
- Wang J, Duncan D, Shi Z, et al. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013;41:W77-83. [Crossref] [PubMed]
- Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013;29:1830-1. [Crossref] [PubMed]
- Huang WY, Hsu SD, Huang HY, et al. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res 2015;43:D856-61. [Crossref] [PubMed]
- Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018;10:277-88. [Crossref] [PubMed]
- Wang X, Xu Z, Chen X, et al. A tropomyosin receptor kinase family protein, NTRK2 is a potential predictive biomarker for lung adenocarcinoma. PeerJ 2019;7:e7125. [Crossref] [PubMed]
- Chen X, Xu Z, Zeng S, et al. SIRT5 downregulation is associated with poor prognosis in glioblastoma. Cancer Biomark 2019;24:449-59. [Crossref] [PubMed]
- Zhou S, Yan Y, Chen X, et al. Roles of highly expressed PAICS in lung adenocarcinoma. Gene 2019;692:1-8. [Crossref] [PubMed]
- Çeliktas M, Tanaka I, Tripathi SC, et al. Role of CPS1 in Cell Growth, Metabolism and Prognosis in LKB1-Inactivated Lung Adenocarcinoma. J Natl Cancer Inst 2017;109:1-9. [Crossref] [PubMed]
- Diez-Fernandez C, Haberle J. Targeting CPS1 in the treatment of Carbamoyl phosphate synthetase 1 (CPS1) deficiency, a urea cycle disorder. Expert Opin Ther Targets 2017;21:391-9. [Crossref] [PubMed]
- Yang C, Fu R, Zhuang Z, et al. Studies on the biological functions of CPS1 in AFB1 induced hepatocarcinogenesis. Gene 2016;591:255-61. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41.e23. [Crossref]
- Yu H, Lee H, Herrmann A, et al. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 2014;14:736-46. [Crossref] [PubMed]
- Teixeira VH, Pipinikas CP, Pennycuick A, et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat Med 2019;25:517-25. [Crossref] [PubMed]
- Oshima G, Poli EC, Bolt MJ, et al. DNA Methylation Controls Metastasis-Suppressive 14q32-Encoded miRNAs. Cancer Res 2019;79:650-62. [Crossref] [PubMed]
- Shahabi S, Kumaran V, Castillo J, et al. LINC00261 Is an Epigenetically Regulated Tumor Suppressor Essential for Activation of the DNA Damage Response. Cancer Res 2019;79:3050-62. [Crossref] [PubMed]


