
Epigenetic plasticity in metastatic dormancy: mechanisms and therapeutic implications
Introduction
Cancer is one of the leading causes of premature mortality across the globe. In a given year, over 18 million people will be diagnosed with cancer, with nearly 10 million people succumbing to their disease (1). The vast majority of these deaths can be directly attributed to metastasis, the process by which cancer cells escape from their primary tumor of origin and disseminate to distant sites (2,3). The deadly nature of metastasis encompasses nearly all tumor types, including breast (4), lung (5), and melanoma (5), as well as colorectal and other gastrointestinal (GI) malignancies (6,7). In spite of this clinical imperative, research into the mechanisms governing metastasis remains under-resourced. Consequently, there is a paucity of therapies capable of specifically targeting metastasis, thereby compelling patients afflicted with metastatic disease to be subjected to cytotoxic chemotherapies (8). Given this tremendous public health burden, there is indeed a pressing need for science and medicine to unravel the molecular mysteries of metastasis.
Further complicating the management and overall outlook for metastatic cancer patients is the emergence of recurrent metastatic disease, which can occur even in the absence of a new primary tumor. Indeed, across the spectrum of human cancers, a substantial proportion of metastatic disease presents years-to-decades following initial diagnosis and treatment (9,10). This delay in the metastatic outgrowth of disseminated cancer cells (DCCs) that reside in metastatic niches is termed metastatic dormancy, a phenomenon encompassing two distinct stages, namely intrinsic (i.e., cellular) and extrinsic (i.e., microenvironmental) dormancy (11). In intrinsic dormancy, DCCs arrest in G0 in response to mitogens, growth factors, and microenvironmental cues to which these cells were previously unresponsive and naïve (12-16). The imposition of such conditions activates prosurvival and stress response mechanisms in dormant DCCs (17,18). In contrast, extrinsic dormancy is defined by the formation of micrometastases by DCCs whose rates of proliferation and apoptosis are roughly equivalent (11). Exit from G0 is accomplished as DCCs adapt to their new microenvironments. However, these cells remain susceptible to immune surveillance (19) and may be reliant upon an insufficient vascular supply (20), thereby limiting their ability to give rise to overt metastatic lesions. Overcoming both intrinsic and extrinsic dormancy requires DCCs to home to and continually remodel specific niches within the metastatic microenvironment (21). DCCs may also co-opt preexisting niches that generate resident tissue stem cells (22). In either case, DCCs must integrate diverse signals arising from parenchymal, stromal, immune, and vascular cells in order to thrive at particular metastatic sites.
DCCs can develop the ability to respond to this milieu of signals through traditional evolutionary processes or via epigenetic control mechanisms. Emerging evidence indicates that DCCs spread early from their primary tumor of origin and undergo limited genetic divergence in metastatic niches (23-25). Thus, the epigenome serves as a critical platform for the acquisition of dormant phenotypes. Herein we summarize major epigenetic regulatory mechanisms and their roles in flexibly maintaining cell identity and dictating metastatic cell behavior, as well as the prospects for targeting these mechanisms therapeutically.
The dynamics of metastatic dormancy
Individual cancer cells do not simultaneously possess all the characteristics required for their dissemination and metastatic outgrowth. Rather, these cells maintain a degree of plasticity that permits them to adopt features that are important at different stages of the metastatic cascade (26-28). Similarly, dormancy requires that DCCs residing within metastatic niches be poised to adapt to the challenges imposed by foreign microenvironments, doing so by dynamically responding to and altering their internal and external states (29,30). Faced with the selective pressures that accompany the metastatic cascade, DCCs do undergo natural selection, which enriches for cells that are well-suited to survive in specific niches at sites of dissemination (31). However, the evolutionary mechanisms that underlie DCC adaptation are limited, particularly in the context of dormancy. There exists a fundamental conflict between quiescence (i.e., cellular dormancy) and proliferation, which is essential for the propagation of de novo mutations. Indeed, quiescence serves as a protective mechanism against genome instability, which ordinarily drives tumor evolution (32,33). Although evolutionary forces may play a more substantial part in extrinsic dormancy (34), micrometastases frequently exhibit significant genetic overlap with their corresponding primary tumors (35,36). Additionally, dormancy requires extensive crosstalk between DCCs and the cells that comprise their microenvironments, including endothelial cells, tissue-resident and circulating immune cells, mesenchymal stem cells, and stromal fibroblasts (37). In general, these cells are viewed as poor substrates for co-evolution with DCCs (38). Hence, Darwinian forces alone are insufficient to give rise to the phenotypic plasticity that is inherent to dormancy.
The tumor-initiating capability of dormant DCCs is intimately linked to their acquisition of cancer stem cell (CSC) traits (39). Dormant DCCs display transcriptional and phenotypic features of CSCs across multiple cancer types (40). Conversely, CSCs are sustained within metastatic niches via signaling pathways that also activate dormancy programs, including Wnt, Notch (39), bone morphogenetic proteins [BMPs; (14,41)], Hedgehog (42), and transforming growth factor-β [TGF-β; (43)]. Several of these pathways converge on common intracellular effector molecules, such as the mammalian target of rapamycin [mTOR; (44)] and the mitogen-activated protein kinases (MAPKs) ERK and p38 (12,45). Ligands that modulate flux through these pathways may be the product of cell-cell communication between DCCs (46); they are also derived from stromal cells (14,41,47), immune cells (19,48), or the extracellular matrix (38,49). Given the distributive nature of these inputs and the need to respond to intrinsic and environmental cues in real time, dormant DCCs must mobilize epigenetic effectors in order to persist indefinitely, retain their self-renewal capacity, and preserve their ability to assume new phenotypes, as we presently outline.
Epigenetic regulation of metastatic dormancy
Several mechanisms for epigenetic regulation of gene expression exist in DCCs. These processes exert their influence at the transcriptional, posttranscriptional, translational, and posttranslational stages of gene expression. In turn, each of these mechanisms possesses unique characteristics that ensure high-fidelity control of cell identity and phenotypic plasticity across the events of intrinsic and extrinsic dormancy.
Covalent modification of DNA and histones
Nucleosomes are composed of genomic DNA that packed around histones, thereby representing the basic units for regulating chromatin accessibility and gene transcription. Covalent modification of the DNA or histone components of nucleosomes at specific genomic loci can dramatically alter gene expression and cellular phenotypes (Figure 1). In dormant DCCs, a transcriptional network responsible for choreographing the G1/S transition is readily suppressed by the DNA methyltransferase DNMT1, instituting cell cycle arrest and quiescence (50). Furthermore, methylation of histone H3 at key residues (H3K4, H3K9, H3K27) results in diminished expression of growth-promoting and CSC-related genes, such as SOX9. In turn, H3 methylation is orchestrated by the orphan nuclear receptor NR2F1, which is commonly epigenetically silenced in multiple cancers via promoter hypermethylation but becomes highly expressed in models of dormancy (51). Similarly, methylation of histone H4 by the histone methyltransferase SMYD5 is required for dormancy in breast DCCs that have disseminated to the lungs (52). These data reveal the versatile nature of nucleosome methylation in coordinating transcriptional programs that are central to establishing dormant and CSC-like identities in DCCs.
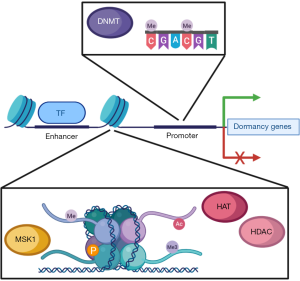
Histone modification takes a number of forms in addition to methylation, and many of these play critical roles in regulating dormancy (Figure 1). For instance, NR2F1 and retinoic acid receptor β (RARβ) act in concert to direct the removal of acetyl groups from histone H3 by histone deacetylases (HDACs), and this axis is associated with the presence of dormant DCCs in patients (51,53,54). Histone acetylation also appears to drive the emergence of a growth-arrested cell population during the development of therapeutic resistance and disease recurrence in glioblastoma (55). Apart from this, the mitogen- and stress-activated protein kinase 1 (MSK1), acting downstream of p38, controls the expression of markers of stemness and differentiation by phosphorylating histone H3 at serine 10 or serine 28 (56). In addition to posttranslational modification, proteolytic processing of a variant histone H3 (H3.3) provokes quiescence concomitant with increased production of TGF-β2 and BMP4 (57). Thus, DCC plasticity is profoundly intertwined with chromatin structure and nucleosome modification, and specific chromatin states program these cells to engage other DCCs and their microenvironments to regulate metastatic outgrowth.
RNA processing
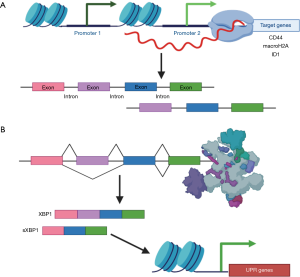
In dormancy, epigenetic regulation via chromatin remodeling not only influences gene expression globally, but also impacts the expression of alternative RNA isoforms in particular (Figure 2A). For example, chromatin structure dictates isoform expression of the CSC surface marker CD44 (58). In addition, upregulation of specific isoforms of the histone macroH2A (macroH2A1.1 and macroH2A2) is indicative of a dormant state and can act as a biomarker for lung cancer recurrence (59). Similarly, the inhibitor of differentiation 1 (ID1) isoform ID1b promotes dormancy by inducing cell cycle arrest at G1/G0 both in vitro and in vivo (60). ID1b represses ERK activation and simultaneously increases the expression of the tumor suppressor p27 and CSC markers NOTCH1 and ALDH1A1. In contrast, the ID1a isoform promotes proliferation, thereby antagonizing the function of ID1b. Interestingly, while the majority of transcript variants associated with dormancy appear to be the result of chromatin remodeling, some posttranscriptional splicing events generate RNAs that directly regulate chromatin architecture (Figure 2B). For instance, the X-box binding protein 1 (XBP1) isoform sXBP-1 activates the unfolded protein response (UPR) in response to cellular stressors typically faced by dormant DCCs, such as hypoxia. sXBP1 is upregulated in CSCs and dormant DCCs, likely because the UPR transcriptional program allows these cells to withstand the stresses imposed by new micrometastatic niches (61,62). RNA processing therefore plays a central role in acclimation and survival of dormant DCCs. Moreover, RNA isoform expression can be interrogated in order to illuminate the mechanisms underlying metastatic recurrence.
Noncoding RNAs (ncRNAs)
ncRNAs are a class of transcribed RNAs that are not translated into proteins. In the past decade, a wealth of important discoveries has established a myriad of functions for both short and long ncRNAs (63-65). For instance, an important class of short ncRNAs are microRNAs (miRNAs), which are 20–24 nucleotides long and base-pair with target mRNAs to either inhibit their translation or induce their cleavage by the RNA-induced silencing complex [RISC; (65)]. miRNAs can function within DCCs or be carried by cancer cell-derived exosomes to repress gene expression by cells of the tumor microenvironment (66,67) (Figure 3). In fact, the abundance of circulating miRNAs, such as miR-222/223, miR-23b, miR-21, miR-34a, miR-127, and miR-197, is correlated with disease progression and dormancy (68-71). The targets of these miRNAs include the chemokine CXCL12, which possesses immunomodulatory functions and controls cancer cell proliferation (71). Similarly, miR-190 is upregulated in dormancy models of glioblastoma, osteosarcoma, liposarcoma, and breast cancer and acts primarily by limiting neoangiogenesis (72,73). Conversely, miR-101 expression is restricted to GI CSCs that give rise to aggressive metastatic disease, rather than those that have adopted a dormant phenotype. A key miR-101 target in mediating its pro-metastatic effects is the histone methyltransferase EZH2, presenting an intriguing intersection between miRNA regulation and chromatin modification in the context of DCC dormancy (74). Taken together, these findings highlight the importance of miRNAs in both intrinsic and extrinsic dormancy at each phase of gene expression.
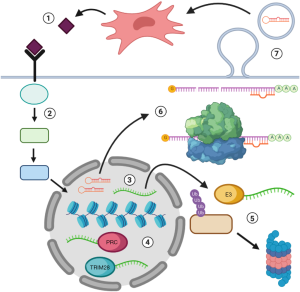
Long non-coding RNAs (lncRNAs) are defined as ncRNAs that are longer than 200 base-pairs and lack an open reading frame; they also have diverse functions in enhancing or suppressing gene expression. In metastatic disease, lncRNAs often carry clinical and functional significance in driving disease progression and recurrence (Figure 3). For example, the lncRNA HOTAIR is a strong predictor for breast cancer metastasis and is correlated with recurrence in cancers of the liver, stomach, bladder, and cervix (75-78). At a molecular level, HOTAIR recruits the polycomb repressive complex (PRC) to specific regions of chromatin, thereby coordinating the epigenetic silencing of target genes such as p21 and Wnt inhibitory factor 1 [WIF1; (79,80)]. Along these lines, expression of the lncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is associated with tumor recurrence and poor prognosis in breast and prostate cancers (81). Like HOTAIR, MALAT1 can assemble the PRC to facilitate histone methylation and control the expression of pro-tumorigenic factors (81,82). The lncRNA BORG (BMP/OP-responsive gene) similarly stimulates metastatic outgrowth in dormant breast DCCs by directing transcriptional repression by the E3 SUMO ligase TRIM28 (83,84). In short, lncRNAs function as primary mediators of the chromatin and transcriptomic alterations that effect metastatic dormancy and reactivation.
In addition to orchestrating transcriptional events, lncRNAs serve to modulate DCC plasticity at the DNA and protein levels (Figure 3). For instance, HOTAIR interacts with the E3 ubiquitin ligases DZIP3 and MEX3B to mark target proteins for proteasomal degradation, particularly in the context of cell cycle arrest (85). On the other hand, BORG promotes the survival of metastasis-initiating cells in response to chemotherapy, a potent inducer of dormancy in discrete subpopulations of DCCs. Specifically, BORG ensures genome stability by coordinating a treatment-induced DNA damage response through its interaction with replication protein A [RPA; (86)]. Still other lncRNAs, such as nuclear paraspeckle assembly transcript 1 [NEAT1; (87)], are associated with tumor recurrence through mechanisms that remain to be fully elucidated. Thus, ncRNAs comprise a vast, multifunctional network that regulates phenotypic plasticity in DCCs through an array of modalities, many of which have yet to be elucidated.
Targeting the epigenome in recurrent and metastatic disease
Because epigenetic mechanisms are paramount in dormancy, they present attractive targets for the development of novel therapeutics against recurrent and metastatic disease. Along these lines, several FDA-approved and commercially available DMNT and HDAC inhibitors have shown promise in preclinical and clinical trials in eradicating CSCs (88), and in improving survival in patients with relapsed and refractory cancers (89). Similarly, DNMT inhibitors in conjunction with poly(ADP-ribose) polymerase (PARP) inhibitors yield clinical benefit in recurrent and resistant breast, ovarian, and urothelial cancers (90,91). Moreover, the DNMT inhibitor azacytidine in combination with the HDAC inhibitor benzamidine demonstrates prolonged progression-free survival in non-small cell lung cancer (NSCLC) patients (92). Trials using DNMT/HDAC inhibitor combinations for metastatic and/or recurrent NSCLC are currently enrolling patients. Ongoing trials are also testing the effects of the HDAC inhibitor belinostat in recurrent B-cell and T-cell lymphomas as well as ovarian cancer (93). Likewise, patients with relapsed multiple myeloma experienced significant survival benefits upon addition of the HDAC inhibitor panobinostat to a treatment regimen containing the proteasome inhibitor bortezomib and dexamethasone (94). Strikingly, next-generation HDAC inhibitors are being designed with high selectivity for specific HDAC isoforms, thereby expanding the promise of these drugs to encompass HDACs that are dysregulated in dormant DCCs (95).
Targeting ncRNAs for the treatment of metastasis presents unique challenges in drug development and delivery. At present, there are no available or experimental therapies that directly target lncRNAs (96). Notably, however, there has been progress made with respect to miRNA therapeutics, particularly those utilizing nanoparticle-conjugated miRNA mimetics to treat multiple solid tumor types (97). This trial represents an exciting advance in the treatment of metastasis, opening the door to synthetic miRNA therapies that preferentially operate in DCCs during dormancy and reactivation. Together with improvements in our molecular knowledge, these novel clinical approaches herald a new age of understanding the epigenome, shedding light on the mechanisms employed by cancer cells to maintain phenotypic plasticity and exploiting these mechanisms to combat the world’s deadliest and most insidious cancers.
Conclusions
Metastatic dormancy is a primary factor underlying disease recurrence and patient mortality, which remain clinically intractable problems in need of innovative therapeutic approaches. Understanding the dynamic interplay between dormancy and the epigenome will give birth to such innovations by advancing our knowledge of the molecular underpinnings of cell plasticity, and by identifying novel targets for the treatment of metastatic cancers. These advances will ultimately facilitate the development of new therapeutic platforms, such as RNA-based therapeutics, leading to improvements in the survival and quality of life for the innumerable people suffering from the burden of metastasis.
Acknowledgments
We thank all members of the Schiemann laboratory for their critical assessment of this Review.
Funding: This work was supported by the National Institutes of Health (CA236273 to WPS; T32GM007250 and F30CA213892 to NJR; and T32GM880316 to KA Parker); and the Case Comprehensive Cancer Center Research Innovation Fund to WPS.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Khalid Sossey-Alaoui) for the Series “Cancer Metastasis: Molecular signaling and therapeutic options” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.02.177). The series “Cancer Metastasis: Molecular signaling and therapeutic options” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell 2017;168:670-91. [Crossref] [PubMed]
- Dillekas H, Rogers MS, Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med 2019;8:5574-6. [Crossref] [PubMed]
- DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin 2019;69:438-51. [Crossref] [PubMed]
- Ascha MS, Ostrom QT, Wright J, et al. Lifetime Occurrence of Brain Metastases Arising from Lung, Breast, and Skin Cancers in the Elderly: A SEER-Medicare Study. Cancer Epidemiol Biomarkers Prev 2019;28:917-25. [Crossref] [PubMed]
- Qiu M, Hu J, Yang D, et al. Pattern of distant metastases in colorectal cancer: a SEER based study. Oncotarget 2015;6:38658-66. [Crossref] [PubMed]
- Sun Z, Zheng H, Yu J, et al. Liver Metastases in Newly Diagnosed Gastric Cancer: A Population-Based Study from SEER. J Cancer 2019;10:2991-3005. [Crossref] [PubMed]
- Sleeman J, Steeg PS. Cancer metastasis as a therapeutic target. Eur J Cancer 2010;46:1177-80. [Crossref] [PubMed]
- Holmgren L, O'Reilly MS, Folkman J. Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat Med 1995;1:149-53. [Crossref] [PubMed]
- Friberg S, Nystrom A. Cancer Metastases: Early Dissemination and Late Recurrences. Cancer Growth Metastasis 2015;8:43-9. [Crossref] [PubMed]
- Yadav AS, Pandey PR, Butti R, et al. The Biology and Therapeutic Implications of Tumor Dormancy and Reactivation. Front Oncol 2018;8:72. [Crossref] [PubMed]
- Sosa MS, Avivar-Valderas A, Bragado P, et al. ERK1/2 and p38alpha/beta signaling in tumor cell quiescence: opportunities to control dormant residual disease. Clin Cancer Res 2011;17:5850-7. [Crossref] [PubMed]
- Ewton DZ, Hu J, Vilenchik M, et al. Inactivation of mirk/dyrk1b kinase targets quiescent pancreatic cancer cells. Mol Cancer Ther 2011;10:2104-14. [Crossref] [PubMed]
- Kobayashi A, Okuda H, Xing F, et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J Exp Med 2011;208:2641-55. [Crossref] [PubMed]
- Endo H, Okami J, Okuyama H, et al. The induction of MIG6 under hypoxic conditions is critical for dormancy in primary cultured lung cancer cells with activating EGFR mutations. Oncogene 2017;36:2824-34. [Crossref] [PubMed]
- Wang H, Yu C, Gao X, et al. The osteogenic niche promotes early-stage bone colonization of disseminated breast cancer cells. Cancer Cell 2015;27:193-210. [Crossref] [PubMed]
- La Belle Flynn A, Calhoun BC, Sharma A, et al. Autophagy inhibition elicits emergence from metastatic dormancy by inducing and stabilizing Pfkfb3 expression. Nat Commun 2019;10:3668. [Crossref] [PubMed]
- Bartkowiak K, Kwiatkowski M, Buck F, et al. Disseminated Tumor Cells Persist in the Bone Marrow of Breast Cancer Patients through Sustained Activation of the Unfolded Protein Response. Cancer Res 2015;75:5367-77. [Crossref] [PubMed]
- Eyles J, Puaux AL, Wang X, et al. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J Clin Invest 2010;120:2030-9. [Crossref] [PubMed]
- Gao D, Nolan DJ, Mellick AS, et al. Endothelial progenitor cells control the angiogenic switch in mouse lung metastasis. Science 2008;319:195-8. [Crossref] [PubMed]
- Gomis RR, Gawrzak S. Tumor cell dormancy. Mol Oncol 2017;11:62-78. [Crossref] [PubMed]
- Yoneda T. Cellular and molecular basis of preferential metastasis of breast cancer to bone. J Orthop Sci 2000;5:75-81. [Crossref] [PubMed]
- Hu Z, Ding J, Ma Z, et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet 2019;51:1113-22. [Crossref] [PubMed]
- Reeves MQ, Kandyba E, Harris S, et al. Multicolour lineage tracing reveals clonal dynamics of squamous carcinoma evolution from initiation to metastasis. Nat Cell Biol 2018;20:699-709. [Crossref] [PubMed]
- Hosseini H, Obradović MMS, Hoffmann M, et al. Early dissemination seeds metastasis in breast cancer. Nature 2016;540:552-8. [Crossref] [PubMed]
- Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell 2011;147:275-92. [Crossref] [PubMed]
- Scheel C, Onder T, Karnoub A, et al. Adaptation versus selection: the origins of metastatic behavior. Cancer Res 2007;67:11476-9; discussion 11479-80. [Crossref] [PubMed]
- Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature 2013;501:328-37. [Crossref] [PubMed]
- Yang X, Liang X, Zheng M, et al. Cellular Phenotype Plasticity in Cancer Dormancy and Metastasis. Front Oncol 2018;8:505. [Crossref] [PubMed]
- De Angelis ML, Francescangeli F, La Torre F, et al. Stem Cell Plasticity and Dormancy in the Development of Cancer Therapy Resistance. Front Oncol 2019;9:626. [Crossref] [PubMed]
- Robinson NJ, Taylor DJ, Schiemann WP. Stem Cells, Immortality, and The Evolution of Metastatic Properties in Breast Cancer. J Cancer Metastasis Treat 2019;5:39. [PubMed]
- Heldt FS, Barr AR, Cooper S, et al. A comprehensive model for the proliferation-quiescence decision in response to endogenous DNA damage in human cells. Proc Natl Acad Sci U S A 2018;115:2532-7. [Crossref] [PubMed]
- Atsumi Y, Fujimori H, Fukuda H, et al. Onset of quiescence following p53 mediated down-regulation of H2AX in normal cells. PLoS One 2011;6:e23432. [Crossref] [PubMed]
- Schmidt-Kittler O, Ragg T, Daskalakis A, et al. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci U S A 2003;100:7737-42. [Crossref] [PubMed]
- Kraus J, Pantel K, Pinkel D, et al. High-resolution genomic profiling of occult micrometastatic tumor cells. Genes Chromosomes Cancer 2003;36:159-66. [Crossref] [PubMed]
- Magbanua MJM, Rugo HS, Hauranieh L, et al. Genomic and expression profiling reveal molecular heterogeneity of disseminated tumor cells in bone marrow of early breast cancer. NPJ Breast Cancer 2018;4:31. [Crossref] [PubMed]
- Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell 2013;155:750-64. [Crossref] [PubMed]
- Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends Genet 2009;25:30-8. [Crossref] [PubMed]
- Hen O, Barkan D. Dormant disseminated tumor cells and cancer stem/progenitor-like cells: Similarities and opportunities. Semin Cancer Biol 2020;60:157-65. [Crossref] [PubMed]
- Weidenfeld K, Barkan D. EMT and Stemness in Tumor Dormancy and Outgrowth: Are They Intertwined Processes? Front Oncol 2018;8:381. [Crossref] [PubMed]
- Gao H, Chakraborty G, Lee-Lim AP, et al. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell 2012;150:764-79. [Crossref] [PubMed]
- Liu S, Dontu G, Mantle ID, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res 2006;66:6063-71. [Crossref] [PubMed]
- Wendt MK, Taylor MA, Schiemann BJ, et al. Downregulation of epithelial cadherin is required to initiate metastatic outgrowth of breast cancer. Mol Biol Cell 2011;22:2423-35. [Crossref] [PubMed]
- Schewe DM, Aguirre-Ghiso JA. ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc Natl Acad Sci U S A 2008;105:10519-24. [Crossref] [PubMed]
- Aguirre-Ghiso JA, Estrada Y, Liu D, et al. ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK). Cancer Res 2003;63:1684-95. [PubMed]
- Malladi S, Macalinao DG, Jin X, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell 2016;165:45-60. [Crossref] [PubMed]
- Bragado P, Estrada Y, Parikh F, et al. TGF-beta2 dictates disseminated tumour cell fate in target organs through TGF-beta-RIII and p38alpha/beta signalling. Nat Cell Biol 2013;15:1351-61. [Crossref] [PubMed]
- Zhang XH, Wang Q, Gerald W, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell 2009;16:67-78. [Crossref] [PubMed]
- Barkan D, El Touny LH, Michalowski AM, et al. Metastatic growth from dormant cells induced by a col-I-enriched fibrotic environment. Cancer Res 2010;70:5706-16. [Crossref] [PubMed]
- Adam AP, George A, Schewe D, et al. Computational identification of a p38SAPK-regulated transcription factor network required for tumor cell quiescence. Cancer Res 2009;69:5664-72. [Crossref] [PubMed]
- Sosa MS, Parikh F, Maia AG, et al. NR2F1 controls tumour cell dormancy via SOX9- and RARbeta-driven quiescence programmes. Nat Commun 2015;6:6170. [Crossref] [PubMed]
- Gao H, Chakraborty G, Lee-Lim AP, et al. Forward genetic screens in mice uncover mediators and suppressors of metastatic reactivation. Proc Natl Acad Sci U S A 2014;111:16532-7. [Crossref] [PubMed]
- Suh YA, Lee HY, Virmani A, et al. Loss of retinoic acid receptor beta gene expression is linked to aberrant histone H3 acetylation in lung cancer cell lines. Cancer Res 2002;62:3945-9. [PubMed]
- Borgen E, Rypdal MC, Sosa MS, et al. NR2F1 stratifies dormant disseminated tumor cells in breast cancer patients. Breast Cancer Res 2018;20:120. [Crossref] [PubMed]
- Rabe M, Dumont S, Alvarez-Arenas A, et al. Identification of a transient state during the acquisition of temozolomide resistance in glioblastoma. Cell Death Dis 2020;11:19. [Crossref] [PubMed]
- Gawrzak S, Rinaldi L, Gregorio S, et al. MSK1 regulates luminal cell differentiation and metastatic dormancy in ER(+) breast cancer. Nat Cell Biol 2018;20:211-21. [Crossref] [PubMed]
- Duarte LF, Young AR, Wang Z, et al. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat Commun 2014;5:5210. [Crossref] [PubMed]
- Luco RF, Allo M, Schor IE, et al. Epigenetics in alternative pre-mRNA splicing. Cell 2011;144:16-26. [Crossref] [PubMed]
- Sporn JC, Kustatscher G, Hothorn T, et al. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 2009;28:3423-8. [Crossref] [PubMed]
- Manrique I, Nguewa P, Bleau AM, et al. The inhibitor of differentiation isoform Id1b, generated by alternative splicing, maintains cell quiescence and confers self-renewal and cancer stem cell-like properties. Cancer Lett 2015;356:899-909. [Crossref] [PubMed]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 2003;23:7448-59. [Crossref] [PubMed]
- Senft D, Ronai ZE. Adaptive Stress Responses During Tumor Metastasis and Dormancy. Trends Cancer 2016;2:429-42. [Crossref]
- Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics 2016;14:42-54. [Crossref] [PubMed]
- Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res 2011;90:430-40. [Crossref] [PubMed]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010;11:597-610. [Crossref] [PubMed]
- Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Bliss SA, Sinha G, Sandiford OA, et al. Mesenchymal Stem Cell-Derived Exosomes Stimulate Cycling Quiescence and Early Breast Cancer Dormancy in Bone Marrow. Cancer Res 2016;76:5832-44. [Crossref] [PubMed]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [Crossref] [PubMed]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008;110:13-21. [Crossref] [PubMed]
- Lim PK, Bliss SA, Patel SA, et al. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res 2011;71:1550-60. [Crossref] [PubMed]
- Ono M, Kosaka N, Tominaga N, et al. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci Signal 2014;7:ra63. [Crossref] [PubMed]
- Naumov GN, Bender E, Zurakowski D, et al. A model of human tumor dormancy: an angiogenic switch from the nonangiogenic phenotype. J Natl Cancer Inst 2006;98:316-25. [Crossref] [PubMed]
- Almog N, Briggs C, Beheshti A, et al. Transcriptional changes induced by the tumor dormancy-associated microRNA-190. Transcription 2013;4:177-91. [Crossref] [PubMed]
- Nishikawa S, Dewi DL, Ishii H, et al. Transcriptomic study of dormant gastrointestinal cancer stem cells. Int J Oncol 2012;41:979-84. [Crossref] [PubMed]
- Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med 2015;12:1-9. [PubMed]
- Xu ZY, Yu QM, Du YA, et al. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci 2013;9:587-97. [Crossref] [PubMed]
- Yan TH, Lu SW, Huang YQ, et al. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour Biol 2014;35:10249-57. [Crossref] [PubMed]
- Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011;18:1243-50. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. [Crossref] [PubMed]
- Huang NS, Chi YY, Xue JY, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget 2016;7:37957-65. [Crossref] [PubMed]
- Biswas S, Thomas AA, Chen S, et al. MALAT1: An Epigenetic Regulator of Inflammation in Diabetic Retinopathy. Sci Rep 2018;8:6526. [Crossref] [PubMed]
- Gooding AJ, Parker KA, Valadkhan S, et al. The IncRNA BORG: A novel inducer of TNBC metastasis, chemoresistance, and disease recurrence. J Cancer Metastasis Treat 2019;5:41. [PubMed]
- Gooding AJ, Zhang B, Jahanbani FK, et al. The lncRNA BORG Drives Breast Cancer Metastasis and Disease Recurrence. Sci Rep 2017;7:12698. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Kim J, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 2013;4:2939. [Crossref] [PubMed]
- Gooding AJ, Zhang B, Gunawardane L, et al. The lncRNA BORG facilitates the survival and chemoresistance of triple-negative breast cancers. Oncogene 2019;38:2020-41. [Crossref] [PubMed]
- Li Y, Li Y, Chen W, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget 2015;6:27641-50. [Crossref] [PubMed]
- Tsai HC, Li H, Van Neste L, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 2012;21:430-46. [Crossref] [PubMed]
- Itzykson R, Thepot S, Berthon C, et al. Azacitidine for the treatment of relapsed and refractory AML in older patients. Leuk Res 2015;39:124-30. [Crossref] [PubMed]
- Criscuolo D, Morra F, Giannella R, et al. New combinatorial strategies to improve the PARP inhibitors efficacy in the urothelial bladder Cancer treatment. J Exp Clin Cancer Res 2019;38:91. [Crossref] [PubMed]
- Pulliam N, Fang F, Ozes AR, et al. An Effective Epigenetic-PARP Inhibitor Combination Therapy for Breast and Ovarian Cancers Independent of BRCA Mutations. Clin Cancer Res 2018;24:3163-75. [Crossref] [PubMed]
- Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 2011;1:598-607. [Crossref] [PubMed]
- Campbell P, Thomas CM. Belinostat for the treatment of relapsed or refractory peripheral T-cell lymphoma. J Oncol Pharm Pract 2017;23:143-7. [Crossref] [PubMed]
- Greig SL. Panobinostat: A Review in Relapsed or Refractory Multiple Myeloma. Target Oncol 2016;11:107-14. [Crossref] [PubMed]
- Roche J, Bertrand P. Inside HDACs with more selective HDAC inhibitors. Eur J Med Chem 2016;121:451-83. [Crossref] [PubMed]
- Blokhin I, Khorkova O, Hsiao J, et al. Developments in lncRNA drug discovery: where are we heading? Expert Opin Drug Discov 2018;13:837-49. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]


