
Donor-derived hypouricemia in irrelevant recipients caused by kidney transplantation
Introduction
Hereditary renal hypouricemia (HRH) is a hereditary and heterogenetic disorder characterized by defective tubular uric acid (UA) transport, reabsorption insufficiency, and/or increased renal urate clearance resulting from the loss-of-function mutations in UA transport genes (1). HRH patients are mostly asymptomatic but 10% of the patients are susceptible to exercise-induced acute renal failure (EIARF) and/or nephrolithiasis, while 20% of them are afflicted by hypercalciuria, which can lead to nephrocalcinosis in the distal tubules (2-4). Currently, two biochemical parameters are in use to diagnose HRH: (I) serum UA concentration less than 2 mg/dL (equivalent to 119 µmol/L), and (II) more than 10% fractional excretion of UA (5,6). In Japan, the rate of HRH incidence is reportedly about 2.54% among hospitalized patients and 0.12–0.72% in the general population (7-9).
The first successful genetically matched kidney transplantation for HRH was reported in 2006 (10). In 2016, a kidney transplant recipient patients with HRH reported a rare case of nephrocalcinosis in the distal tubules three months after transplant surgery (4). Other than these sporadic reports, there is a dearth of scientific data on the transmission of renal hypouricemia in irrelevant donor-recipient transplantation.
Over the past decade, genome-wide association studies and case reports have shown an increase in the number of genetic variants that influence serum UA concentrations, such as SLC2A9, SLC22A12, SLC17A3, and ABCG2 (11,12). Single nucleotide polymorphisms (SNPs) in regulatory regions (rSNPs) modulate levels of gene expression in an allele-specific manner; however, there is lack of such studies in kidney transplantation research. Further, majority of published studies on hypouricemia includes case report or case series, which lack essential statistical analysis and comparison with healthy controls. Herein, we present two unrelated recipients who had no history of hypouricemia before renal transplantation but experienced sudden and unexpected hypouricemia after receiving transplants from a donor of different genetic background. We performed a DNA sequencing analysis in one healthy control and two kidney transplant recipients and followed up for 3.5 years after their surgery. Our results showed differential gene expression profile between healthy individuals and HRH patients and indicated possible pathogenic pathways associated with disease onset and progression.
Methods
Biochemical and ultrasound evaluation
To evaluate the factors associated with hypouricemia, we checked parameters such as blood biochemistry and urine routine for the UA metabolism, renal tubular acidosis and urine electrolytes, and the liver was examined by ultrasound.
Tissue samples
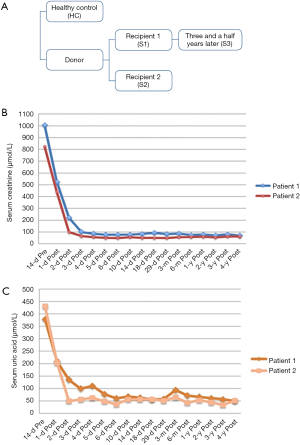
Two of the three transplant specimens were collected at the time of transplantation and preserved at the hospital, while the other transplant specimen was collected in recipient 1 followed up for 3.5 years after transplantation surgery. Renal biopsy tissue was obtained from a live healthy renal transplant-recipient and the sample was used as the healthy control (Figure 1A). The study details were explained to all the participants, and a signed informed consent was obtained after their agreement. Extraction of DNA was performed using the Axyprep™ Blood Genomic DNA Miniprep Kit (Axyprep, USA) following the manufacturer’s recommendations. DNA was eluted in approximately 100 µL of buffer AE. DNA integrity was checked on 1% agarose gel and purity were checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). DNA was quantified using Qubit® DNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA).
Library preparation and sequencing
A total of 700 ng DNA from each sample was used as the input material for the DNA library preparations. Sequencing libraries were generated using NEB Next® Ultra DNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. The NEB Next Adaptor with hairpin loop structure were ligated to 3' adenylated DNA fragments to prepare for hybridization and electrophoresis was carried out to select DNA fragments of specified length. Subsequently, 3 µL USER Enzyme (NEB, USA) was used with size-selected DNA at 37 °C for 15 min and 95 °C 5 min before carrying out polymerase chain reaction (PCR). PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer to enrich final adaptor modified fragmented sample. Finally, the library fragments were purified using AMPure XP system (Beckman Coulter, Beverly, USA). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using HiSeq 2500 PE Cluster Kit (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform.
PCR amplification and sequence analysis
The genomic DNA was isolated from the transplant samples obtained from the recipients and live donors using the AxyprepTM Blood Genomic DNA Miniprep Kit (Axyprep, USA). Seven pairs of oligonucleotide primers were generated to amplify the different regions obtained from DNA sequencing and were sequenced directly. A total of 80 ng of genomic DNA was amplified in 20 µL reaction volume containing 10 µL Premix Taq (TaKaRa) and 0.8 µM primers. Amplification products were purified on 1.5% agarose gel using 0.5× TBE buffer and Wizard SV gel and Gel/PCR DNA Fragments Extraction Kit (Promega, USA). DNA sequencing was performed with an automated DNA sequencer (Applied Biosystems 3730-Avant Genetic Analyzer; Applied Biosystems, USA).
RNA isolation and cDNA library construction
Total RNA was extracted from implants of two kidney transplant recipient using Trizol reagent (Invitrogen, USA), and RNase-free DNase I (TaKaRa, Japan) following the manufacturer’s protocol. One was the healthy control and the other was recipient1 after three and a half years post-transplantation. A total of 1.5 µg of RNA per sample was used as input material for RNA sample reparations. The differentially expressed genes were detected using an Affymetrix Mouse Genome 430 2.0 microarray (Thermo Fisher Scientific). The experimental procedures for microarray were performed at the Hangzhou Tianke Corporation (Hangzhou, China). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq SR Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After the cluster generation, the library preparations were sequenced on an Illumina HiSeq 2500 platform.
Results
Clinical and biochemical investigations
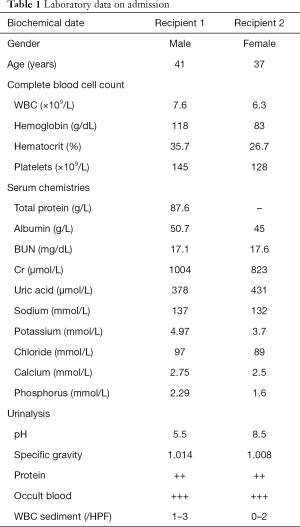
On September 2012, a 41-year-old male (recipient 1) and a 37-year-old female (recipient 2) with end-stage renal disease caused by chronic kidney disease (CKD) received renal transplantation from a deceased donor who had died from craniocerebral injury. On admission, they had a serum creatinine of 1,004 and 823 µmol/L (Table 1). Recipient 1 had a gradually increased serum creatinine (S-Cr) level beginning in 2000 diagnosed as IgA nephritis and started on hemodialysis at the age of 34 years. Serum creatinine level gradually increased in recipient 2 for 8 years and she began hemodialysis at the age of 35 years. The donor was a 30-year-old male (serum creatinine: 79 µmol/L) with no significant past medical history (Figure 1A). Within the first week of transplant, the serum creatinine levels decreased to 76 and 49 µmol/L respectively (Figure 1B). In addition, their physical examination, laboratory examination and grafts biopsy at zero time were uneventful and they were released from the hospital after great recovery. Both of them received triple immunosuppressive therapy consisting of cortico-steroids, mycophenolate mofetil, and tacrolimus. Since then, the two recipients have reported fluctuating low serum UA levels (patient 1: 55–65 µmol/L, normal serum creatinine: 70–80 µmol/L; patient 2: 45–55 µmol/L, normal serum creatinine 60–70 µmol/L) (Figure 1C). We confirmed a well-functioning kidney graft (serum creatinine 82 µmol/L, eGFR 102 mL/min and serum creatinine 60 µmol/L, eGFR 109 mL/min respectively) with no proteinuria or haematuria after three years the transplantation except for the high fraction excretion of UA (FEUA) of 44% and 75% (normal <10%) and UA clearance of 35.9 and 73.3 mL/min (normal 7.3–14.7 mL/min). We reviewed the results of pre-operative laboratory examinations of donor and found that the donor had a very low serum UA of 48 µmol/L. Our findings suggested that the low serum UA in two recipients could be associated with HRH, and therefore, molecular genetic analysis was performed to confirm the same.
Full table
UA transporter genes analysis
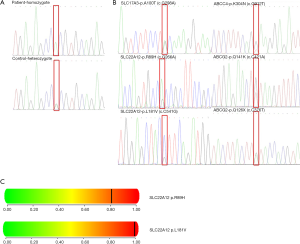
PCR followed by DNA sequence analysis revealed 7 types of mutations. Probands were heterozygous for the unpublished missense mutation p.Q141K(c.C421A) in exon 5 and p.Q126X(c.C376T) in exon 4 in the ABCG2 and heterozygous for the unidentified missense mutation p.R89H(c.G266A) and p.L181V(c.C541G) in exon 1 in the SLC22A12 gene. Variants p.R89H and p.L181V are novel and have not yet been identified in SLC22A12 gene. Moreover, the nature of these mutations appears pathogenic as per the PolyPhen software (http://genetics.bwh.harvard.edu/pph2/) indicates that substitutions in SLC22A12 were probably damaging (score of 0.809; sensitivity 0.84; specificity 0.93 and score of 0.996; sensitivity 0.55; specificity 0.98, respectively).
Other variations, one homozygous exon variant (p.R294H) and one heterozygous exon variant (p.A100T) have been previously reported (Figure 2, Table 2).

Full table
Gene expression analysis by RNA-sequencing (RNA-seq)
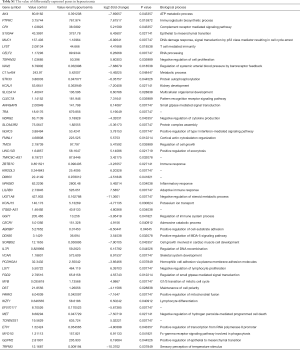
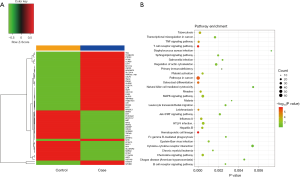
To study the effect of mutations on the gene expression, we analyzed the transcriptomes of the transplant tissues by RNA-seq (Table S1). Analysis of the RNA-seq data revealed that a total of 57 genes were differentially regulated among the hypouricemia patients and the healthy controls (fold change >2, P value <0.05). Out of 57 gene, 21 were upregulated, while 36 genes were down regulated. We used unsupervised clustering hierarchy (Figure 3A) and the details of the differentially expressed genes are given in Table 3. KEGG pathway analysis revealed that the differentially expressed genes were played roles in hematopoietic cell lineage, T cell receptor signaling pathway, cancer related pathways, MAPK signaling pathway, and other important regulatory processes (Figure 3B).
Full table

Full table
Relationship between gene mutation and expression levels
We tried to elucidate the relationship between SNPs, gene expression, and phenotypes together. Mutation analysis findings point towards epithelial growth factor (EGF) receptor (EGFR), IL-7 receptor (IL7R) and growth hormone receptor (GHR) which could implicate the important role in transcriptional regulation through cancer related pathways, MAPK signaling pathway, regulation of actin cytoskeleton cytokine-cytokine receptor interaction, hematopoietic cell lineage, and Jak-STAT signaling pathway to exert their influence on the phenotypes (Table 3).
Discussion
HRH, is defined arbitrarily as serum UA concentration less than 119 µmol/L and increased fractional excretion of uric acid (FEUA) and/or uric acid clearance (CUA), with exclusion of other diseases that present hypouricemia as a symptom (13). Loss-of-function mutations in the SLC22A12 gene coding the UA transporter 1 (URAT1) and SLC2A9 gene coding the glucose transporter (GLUT9) caused type 1 (RHUC1) and 2 (RHUC2), respectively. Most renal hypouricemia is caused by mutations in the SLC22A12 gene. The high incidence of RHUC1 has been reported in the Asia region and Roma ethnicity. The allele frequency of c.774G>A (p.W258X) and c.269G>A (p.R90H) were 2.37% and 0.40% in SLC22A12 among Japanese and Koreans (14,15). Frequencies of the c.1245_1253del and c.1400C>T variants were present in the Roma population at 1.87% and 5.56%, respectively (16,17).Several GWAS have indicated a substantial association between urate concentration and SNPs at 10 genetic loci including transporter-coding genes such as SLC2A9 (GLUT9), ABCG2 (BCRP), SLC17A1 (NPT1), SLC17A3 (NPT4), SLC17A4 (provisionally named as NPT5), SLC22A11 (OAT4), SLC22A12 (URAT1), and SLC16A9 (MCT9) as well as urate transport related scaffolding protein PDZK1 (18). However, Hurba et al. (19) reported the non-synonymous allelic variants on of GLUT9 were not related to urate uptake activity. But several studies reported clear function impact of GLUT9 variants in patients with renal hypouricemia 2 (20-22). For example, Dinour et al. (23) reported that homozygous mutations of GLUT9 cause a total defect of UA absorption and are associated with a high incidence of renal calculus and EIAKI and nephrolithiasis. Previously, a successful living-related kidney transplant has been reported in HRH. Both the donor and the recipient had the same disorder of urate metabolism and were homozygous for G774A before kidney transplantation (10). Another rare case reported nephrocalcinosis in the distal tubules caused by HRH in a living-donor renal transplantation. Genetic analysis revealed a heterozygous nonsense mutation of C889T in exon 5 of the urate transporter 1 (URAT1) gene in both, the donor and the recipient (4). In this study, we present a rather rare case of donor-derived HRH. To the best of our knowledge, this is the first report to show that unrelated recipients can acquire unexpected hypouricemia after kidney transplantation from the same donor with a different genetic background.
DNA analysis was performed on the tissue before being transplanted into the two recipients. The cases and a control were followed for 3.5 years post-transplantation. Our results showed that the mutated genes in the grafts of the donor remained unchanged after transplantation in a different un-URH environment up to the follow-up duration of 3.5 years. Many non-pathogenic single point mutations identified in the present study have been reported earlier and included the homozygous missense mutation, p.R294H in SLC2A9 in exon 7 (24) and a heterozygous sequence variant, p.A100T in SLC17A3 in exon 3 (25). We could not confirm the nosogenetic mutations from the family of the deceased donor. Therefore, the effect of previously unreported mutations on the hypouricemia remains unknown and needs to be answered in future. Genetic variants have been associated with many human diseases. However, about 88% of the GWAS-nominated SNPs are in intronic or intergenic regions suggesting that the noncoding regions of the genome can contribute to the disease risk, and may be involved in gene regulation. However, the underlying mechanism by remains unclear (26). SNPs can modulate the gene expression through a change in chromatic structure to distance a gene from its enhancers and by altering the copy number (27). Within each susceptibility locus, candidate risk genes have been prioritized based largely on bioinformatic evaluations of the relationships among genes, the presence of coding SNPs, or the gene expression-genotype correlations (28-30). Regulatory and coding variants often modify the functional impact of each other that can be detected by the sequencing data. Characterizing these mutual effects might help us understand functional mechanisms behind genetic associations to human phenotypes (31). For example mutational signatures related to liver carcinogenesis revealed frequently mutated coding and noncoding regions, such as long intergenic noncoding RNA genes (NEAT1 and MALAT1), promoters, CTCF-binding sites, and regulatory regions (32). Biswajit et al. unveiled rs2279590 at PTK2B–CLU locus, a risk factor previously associated with Alzheimer’s disease, to have an enhancer effect on two nearby genes coding for protein tyrosine kinase 2 beta (PTK2B) and epoxide hydrolase-2 (EPHX2) (33). Based on these results, we speculate that defects in DNA sequences could probably affect tubule function through differential RNA expression. DNA mutation analysis have identified three risk loci that increase renal hypouricemia risk: (I) SNP (rs62452902) at EGFR locus, (II) SNP (rs10058453) at IL7R locus, (III) SNP (rs4146624) at GHR. We performed enrichment analysis from the data of the differentially expressed genes and identified that pathways related to cancers, MAPK signaling, regulation of actin cytoskeleton, cytokine-cytokine receptor interaction, hematopoietic cell lineage, and Jak-STAT signaling were significantly altered in hypouricemia transplant tissue, indicating that these pathway may be involved in the disease. EGF risk alleles may upregulate pathways related to cancer, MAPK signaling, alter actin cytoskeleton, and cytokine-cytokine receptor interactions to promote elevated blood UA by impacting UA metabolism and inhibiting UA excretion. Though our study presents interesting findings, its limitations include small number of cases and therefore requirement of further work to validate this work.
Conclusions
We report a renal transplantation case of donor-derived hypouricemia caused by mutations in the transplant tissue of donor with HRH. Seven kinds of potent mutations were discovered in this case, including two novel mutations which could be pathogenic in nature. However, further studies are needed to prove the role of these mutations in HRH pathogenicity.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (No. 81970647).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study details were explained to all the participants, and a signed informed consent was obtained after their agreement.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Stiburkova B, Sebesta I, Ichida K, et al. Novel allelic variants and evidence for a prevalent mutation in URAT1 causing renal hypouricemia: biochemical, genetics and functional analysis. Eur J Hum Genet 2013;21:1067-73. [Crossref] [PubMed]
- Ichida K, Hosoyamada M, Hisatome I, et al. Clinical and molecular analysis of patients with renal hypouricemia in Japan-influence of URAT1 gene on urinary urate excretion. J Am Soc Nephrol 2004;15:164-73. [Crossref] [PubMed]
- Ochi A, Takei T, Ichikawa A, et al. A case of acute renal failure after exercise with renal hypouricemia demonstrated compound heterozygous mutations of uric acid transporter 1. Clin Exp Nephrol 2012;16:316-9. [Crossref] [PubMed]
- Okabayashi Y, Yamamoto I, Komatsuzaki Y, et al. Rare case of nephrocalcinosis in the distal tubules caused by hereditary renal hypouricaemia 3 months after kidney transplantation. Nephrology (Carlton) 2016;21 Suppl 1:67-71. [Crossref] [PubMed]
- Sebesta I, Stiburkova B, Bartl J, et al. Diagnostic tests for primary renal hypouricemia. Nucleosides Nucleotides Nucleic Acids 2011;30:1112-6. [Crossref] [PubMed]
- Shen H, Feng C, Jin X, et al. Recurrent exercise-induced acute kidney injury by idiopathic renal hypouricemia with a novel mutation in the SLC2A9 gene and literature review. BMC Pediatr 2014;14:73. [Crossref] [PubMed]
- Hisatome I, Ogino K, Kotake H, et al. Cause of persistent hypouricemia in outpatients. Nephron 1989;51:13-6. [Crossref] [PubMed]
- Takahashi T, Tsuchida S, Oyamada T, et al. Recurrent URAT1 gene mutations and prevalence of renal hypouricemia in Japanese. Pediatr Nephrol 2005;20:576-8. [Crossref] [PubMed]
- Wakasugi M, Kazama JJ, Narita I, et al. Association between hypouricemia and reduced kidney function: a cross-sectional population-based study in Japan. Am J Nephrol 2015;41:138-46. [Crossref] [PubMed]
- Yamamoto I, Yamamoto H, Ichida K, et al. Successful living-related kidney transplantation in hereditary renal hypouricaemia. Nephrol Dial Transplant 2006;21:2041. [Crossref] [PubMed]
- Enomoto A, Kimura H, Chairoungdua A, et al. Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature 2002;417:447-52. [Crossref] [PubMed]
- Dehghan A, Köttgen A, Yang Q, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet 2008;372:1953-61. [Crossref] [PubMed]
- Nakayama A, Matsuo H, Ohtahara A, et al. Clinical practice guideline for renal hypouricemia (1st edition). Hum Cell 2019;32:83-7.
- Iwai N, Mino Y, Hosoyamada M, et al. A high prevalence of renal hypouricemia caused by inactive SLC22A12 in Japanese. Kidney Int 2004;66:935-44. [Crossref] [PubMed]
- Cheong HI, Kang JH, Lee JH, et al. Mutational analysis of idiopathic renal hypouricemia in Korea. Pediatr Nephrol 2005;20:886-90. [Crossref] [PubMed]
- Stiburkova B, Gabrikova D, Čepek P, et al. Prevalence of URAT1 allelic variants in the Roma population. Nucleosides Nucleotides Nucleic Acids 2016;35:529-35. [Crossref] [PubMed]
- Gabrikova D, Bernasovska J, Sokolova J, et al. High frequency of SLC22A12 variants causing renal hypouricemia 1 in the Czech and Slovak Roma population; simple and rapid detection method by allele-specific polymerase chain reaction. Urolithiasis 2015;43:441-5. [Crossref] [PubMed]
- Anzai N, Jutabha P, Amonpatumrat-Takahashi S, et al. Recent advances in renal urate transport: characterization of candidate transporters indicated by genome-wide association studies. Clin Exp Nephrol 2012;16:89-95. [Crossref] [PubMed]
- Hurba O, Mancikova A, Krylov V, et al. Complex analysis of urate transporters SLC2A9, SLC22A12 and functional characterization of non-synonymous allelic variants of GLUT9 in the Czech population: no evidence of effect on hyperuricemia and gout. PLoS One 2014;9:e107902. [Crossref] [PubMed]
- Dinour D, Gray NK, Ganon L, et al. Two novel homozygous SLC2A9 mutations cause renal hypouricemia type 2. Nephrol Dial Transplant 2012;27:1035-41. [Crossref] [PubMed]
- Matsuo H, Chiba T, Nagamori S, et al. Mutations in glucose transporter 9 gene SLC2A9 cause renal hypouricemia. Am J Hum Genet 2008;83:744-51. [Crossref] [PubMed]
- Ruiz A, Gautschi I, Schild L, et al. Human Mutations in SLC2A9 (Glut9) Affect Transport Capacity for Urate. Front Physiol 2018;9:476. [Crossref] [PubMed]
- Dinour D, Gray NK, Campbell S, et al. Homozygous SLC2A9 mutations cause severe renal hypouricemia. J Am Soc Nephrol 2010;21:64-72. [Crossref] [PubMed]
- Stiburkova B, Taylor J, Marinaki AM, et al. Acute kidney injury in two children caused by renal hypouricaemia type 2. Pediatr Nephrol 2012;27:1411-5. [Crossref] [PubMed]
- Stiburkova B, Ichida K, Sebesta I. Novel homozygous insertion in SLC2A9 gene caused renal hypouricemia. Mol Genet Metab 2011;102:430-5. [Crossref] [PubMed]
- Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 2009;106:9362-7. [Crossref] [PubMed]
- Weischenfeldt J, Symmons O, Spitz F, et al. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet 2013;14:125-38. [Crossref] [PubMed]
- Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119-24. [Crossref] [PubMed]
- Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 2011;43:246-52. [Crossref] [PubMed]
- Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet 2008;40:955-62. [Crossref] [PubMed]
- Lappalainen T, Montgomery SB, Nica AC, et al. Epistatic selection between coding and regulatory variation in human evolution and disease. Am J Hum Genet 2011;89:459-63. [Crossref] [PubMed]
- Fujimoto A, Furuta M, Totoki Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet 2016;48:500-9. [Crossref] [PubMed]
- Padhy B, Hayat B, Nanda GG, et al. Pseudoexfoliation and Alzheimer's associated CLU risk variant, rs2279590, lies within an enhancer element and regulates CLU, EPHX2 and PTK2B gene expression. Hum Mol Genet 2017;26:4519-29. [Crossref] [PubMed]


