
In Silico analyses of host immunity and stroma provide prognostic factors in early stage pancreatic ductal adenocarcinoma (PDAC)
Pancreatic ductal adenocarcinoma (PDAC) remains a challenging cancer to treat and with a 5-year overall survival (OS) of approximately 8% in the United States (1). Patients with resectable tumors (Stage I/II) who receive adjuvant therapy have a 5-year overall survival of approximately 30%. Prognosis is dismal for patients with stage IV disease with a median survival of about 12 months. Despite significant advances in the development of immune checkpoint inhibitors (e.g., anti-PD-1, anti-CTLA4) and targeted therapies (e.g., tyrosine kinase inhibitors), these treatment approaches have not resulted in improved outcomes in PDAC. It is well established that the tumor microenvironment (TME) is directly involved in the aggressive behavior of PDAC and contributes to the ineffectiveness of current therapy. As such, further defining the complexities of the TME could play an important role in identifying new prognostic markers that may better stratify patients for novel therapeutics in PDAC.
The TME is comprised of non-tumor cells, including the extracellular matrix (ECM) proteins, pancreatic stellate cells, blood vessels, immune cells, macrophages, and inflammatory cytokines/chemokines. These components directly and/or indirectly interact with the tumor cells and with one another in a complex manner that promotes tumor growth, invasion, and a suppressed host immunity (Figure 1). The PDAC TME is unique, complex, and possesses characteristics that directly impact a gene expression continuum of tumor cells (2). One unique feature of PDAC is fibrosis due to production of excessive stroma known as the ‘desmoplastic reaction’ (3). This dense stromal tissue creates a hypovascular, hypoxic TME milieu which is a physicochemical barrier that impacts drug delivery to cancer cells. Although tumor-stroma interactions contribute to chemotherapy resistance in PDAC, there are other important characteristics of the TME that contribute to its aggressive nature (4).
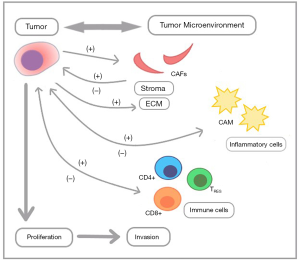
Another important factor of the TME is immune suppression (‘cold’ tumor), although the exact mechanism(s) of resistance to immune checkpoint therapy is not fully understood. Current evidence in PDAC points to limited mutational heterogeneity, low tumor mutational burden, and a lack of neoantigens contributing to low immunogenicity (5). The PDAC TME is infiltrated by sparse immune cells, including regulatory T cells (TREG), cancer associated fibroblasts (CAFs), cancer associated macrophages (CAMs), and myeloid-derived suppressor cells which are cancer associated neutrophils (CANs). Evidence from multiple studies supports the theory that interactions between cancer cells and the cells of the TME affects the heterogeneity, response to therapy, and progression of PDAC. This highlights the need for a more comprehensive understanding of the microenvironmental gene expression signatures in order to define immune cell activity in PDAC (6).
Genomics have confirmed four common recurrent pathogenic mutations in PDAC: KRAS, TP53, CDKN2A, and SMAD4. KRAS is mutated in >90% and TP53 is mutated in approximately 70% of PDACs (5,7). Given the complex pathophysiology of PDAC, there are ongoing efforts to better characterize immune and stromal components in the TME that affect the genomics of cancer cells and have an impact on clinical outcomes. Since the TME has prognostic significance (8,9), Pu et al. analyzed the TCGA database to extend these observations by identifying 147 early stage PDAC cases of which 146 had mutational data, 141 had OS data, and 122 had relapse-free survival (RFS) data (9). The MAlignant Tumor tissues using Expression data (ESTIMATE) algorithm was used to calculate immune and stromal scores. Cell infiltrations of the TME were analyzed by the Tumor IMmune Estimation Resource (TIMER). Of the four common gene mutations, KRAS mutant cases had significantly lower immune and stromal scores, TP53 mutant cases had low immune scores, and CDKN2A and SMAD4 mutant cases were neutral. Given that KRAS and TP53 mutant cases predominate in PDAC, low immune scores confirm previous studies (6). However, there was no difference in OS or RFS for both high versus low immune and stromal score (Pu et al., figure 2), respectively. This is likely due to the analysis of primarily early stage patients (Stage I/II) in this study, which is a minority of PDAC cases.
Innate and adaptive immune cell infiltrations are higher in high immune and stromal score groups, except that CD4+ T-cells were low in the high stroma group and macrophages were low in the high immune group (Pu et al., figure 3). Differentially expressed genes (DEGs) within high versus low immune and stromal score groups indicated many up-regulated genes (n=124) and a small number of down-regulated genes (n=6) that were common to both immune and stromal groups. Top GO terms and KEGG pathways enriched to immune cells (CNR2 module) and cytokines (CCL22 module) identified by STRING and Cytoscape, were of prognostic value. Both CNR2 and CCL22 enrich to CAMs that require further investigation regarding T-cells and tumor cells within the TME (9,10). This study did not capture CA19-9, a prognostic marker that may also help stratify patients to immune/stroma subtypes.
A major limitation of this study is the lack of TME data from patients with advanced disease, with only 5% of the study population having stage III or IV disease. By contrast, more than 80% of cases of PDAC present with regional nodal involvement or metastatic disease (11). Other limitations include the lack of information regarding the cellularity of the tumor and its heterogeneity, contributions of genomic instability to disease heterogeneity, and the issue of low tumor cellularity when compared to stroma (12). In addition, non-genetic mechanisms play a key role and proteomics of the ECM have revealed differential contributions of tumor and cells of the microenvironment to PDAC progression (13). Further classification of PDAC to basal-like and classical-like (12) in addition to epigenetic and metabolomic regulation of gene expression may provide better therapeutic interventions. Despite the aforementioned limitations, this study helps to build upon our current understanding of the characteristics of PDAC microenvironmental gene expression and provides potential directions for further investigation.
Acknowledgments
Funding: We acknowledge the Mays Cancer Center for the NCI CCSG P30 core grant for support.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Kleeff J, Beckhove P, Esposito I, et al. Pancreatic cancer microenvironment. Int J Cancer 2007;121:699-705. [Crossref] [PubMed]
- Neesse A, Michl P, Frese KK, et al. Stromal biology and therapy in pancreatic cancer. Gut 2011;60:861-8. [Crossref] [PubMed]
- Mahadevan D, Von Hoff DD. Tumor-stroma interactions in pancreatic ductal adenocarcinoma. Mol Cancer Ther 2007;6:1186-97. [Crossref] [PubMed]
- Tran E, Ahmadzadeh M, Lu YC, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015;350:1387-90. [Crossref] [PubMed]
- Morrison AH, Byrne KT, Vonderheide RH. Immunotherapy and prevention of pancreatic cancer. Trends Cancer 2018;4:418-28. [Crossref] [PubMed]
- Raphael BJ, Hruban RH, Aguirre AJ, et al. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017;32:185-203.e13. [Crossref] [PubMed]
- Qian ZR, Rubinson DA, Nowak JA, et al. Association of alterations in main driver genes with outcomes of patients with resected pancreatic ductal adenocarcinoma. JAMA Oncol 2018;4:e173420. [Crossref] [PubMed]
- Pu N, Chen Q, Gao S, et al. Genetic landscape of prognostic value in pancreatic ductal adenocarcinoma microenvironment. Ann Transl Med 2019;7:645. [Crossref] [PubMed]
- Mahajan UM, Langhoff E, Goni E, et al. Immune cell and stromal signature associated with progression-free survival of patients with resected pancreatic ductal adenocarcinoma. Gastroenterology 2018;155:1625-39.e2. [Crossref] [PubMed]
- SEER Cancer Stat Facts: Pancreatic Cancer. National Cancer Institute. Bethesda, MD. Available online: https://seer.cancer.gov/statfacts/html/pancreas.html
- Chan-Seng-Yue M, Kim JC, Wilson GW, et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet 2020;52:231-40. [Crossref] [PubMed]
- Tian C, Clauser KR, Ohlund D, et al. Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc Natl Acad Sci U S A 2019;116:19609-18. [Crossref] [PubMed]


