
Development of a prognostic index of colon adenocarcinoma based on immunogenomic landscape analysis
Introduction
According to the Global Cancer Report released at the end of 2018, colon adenocarcinoma (COAD) is one of the most commonly diagnosed cancers, and one of the top three malignant tumors in terms of morbidity and mortality (1). Considering that the early symptoms are not visible, most patients diagnosed with COAD are in the middle or late stages of the disease, which impedes treatment and timely surgical intervention. Therefore, despite the increasing number of targeted drugs (2,3), the 5-year average survival rate of COAD patients in China is only 57%. To improve this rate, the prognosis status of patients should be monitored closely, and the treatment plans should be timely adjusted. This necessitates the development of an independent prognostic index that allows for the evaluation of COAD clinical outcomes.
Evidence of the close relation between the immune microenvironment and tumor development has substantially increased over the past few years (4-7). Consequently, tumor immunotherapy has emerged as a promising technique of treatment, and it has garnered the attention of many researchers (8,9). This technique relies on the activation of immune cells that can trigger anti-tumor immune responses, leading to the elimination of tumor residual lesions, the inhibition of tumor growth, and the breaking of immune tolerance (10). Unfortunately, immunotherapy is not effective for all tumor patients. Therefore, clinical research efforts are currently focused on the identification of patients who can be efficiently treated using immunotherapy techniques. This may be accomplished by determining the type and proportion of immune cells and factors in the tumor immune microenvironment, which are in turn controlled by gene expression. The recent progress in gene sequencing technology has allowed for the design of new and sensitive gene-based biomarkers (11,12) that may be used to improve the prognosis of COAD. The comprehensive analysis of the relevance between the immune-related genes (IRGs) and survival is conducive to elucidating the potential prognostic value of these genes. For example, an immuno-genomic study of tumor immune microenvironment has previously shown that mRNA-seq is effective in predicting clinical response (5).
In this study, we identified the differentially expressed IRGs in COAD patients by relating the gene expression profiles to the IRGs from different online databases. The functional enrichment analysis and prognostic value of the survival-related IRGs were also determined. The prognostic index was finally established based on Cox regression analysis, followed by model evaluation and application. In theory, the robust prognostic biomarker reported herein can be used to predict the clinical outcomes of therapeutic methods, including cancer immunotherapy.
Methods
Acquisition of gene expression data and IRGs
The RNA-seq data of COAD samples was taken from The Cancer Genome Atlas (TCGA) database (13) that is published in the Genomic Data Commons (GDC) portal (https://portal.gdc.cancer.gov/) (14), and the clinical information of these samples was downloaded and extracted by Perl. The collected dataset is comprised of 473 COAD samples and 41 adjacent normal samples. In addition, a comprehensive list of IRGs (1,811 in total) was downloaded from the Immunology Database and Analysis Portal (ImmPort) (https://www.immport.org/home) (15). These genes were divided into 17 categories based on their molecular functions (MFs) (e.g., cytokine, interleukin, and natural killer cell cytotoxicity).
Identification of differentially expressed genes (DEGs)
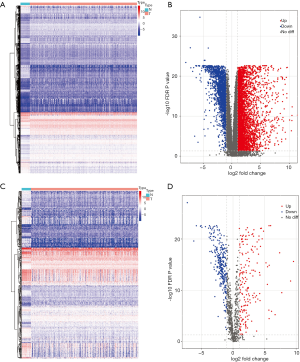
The DEGs were identified by comparing the COAD and adjacent normal samples using R package “limma” (16). Specifically, the genes exhibiting |log2(fold change)| >1 and false discovery rate (FDR) <0.05 were considered as DEGs. Afterwards, the differentially expressed IRGs were extracted from the list of identified DEGs. Heat maps and volcano plots of DEGs and differentially expressed IRGs were generated by R package pheatmap and ggplot2, respectively (17,18).
Screening of survival-related IRGs and evaluation of their prognostic value
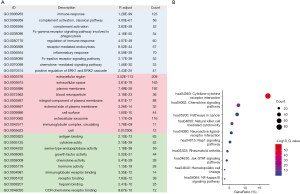
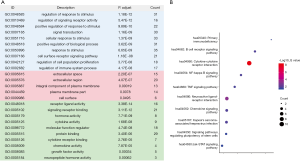
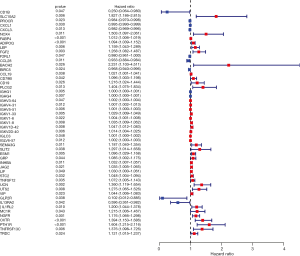
A survival analysis of all of the differentially expressed IRGs was performed using the R package “survival” (19), and the survival-related IRGs were determined by univariate Cox analysis (P<0.05). In order to explore the potential molecular mechanisms of differentially expressed IRGs and survival-related IRGs, functional enrichment analysis were conducted on the basis of Kyoto encyclopedia of genes and genomes (KEGG) and gene ontology (GO) database, including biological process (BP), MF and cellular component (CC) (20,21), using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.8 (22). The GO terms and KEGG signaling pathways were enriched with an adjusted P value of <0.05. Moreover, the top 10 KEGG signaling pathways were drawn into a bubble chart using the R package “ggplot2” (18). To determine the prognostic value of survival-related IRGs, a forest plot of these genes was constructed using the hazard ratio (HR) as an indicator, based on univariate Cox analysis.
Development of the IRG-based prognostic index (IRGPI)
Survival-related IRGs were submitted for multivariate analysis to develop the IRGPI, and integrated IRGs was kept as independent prognostic indicators. Specifically, the IRGPI was established by multiplying the expression values with the Cox regression coefficient (23). It should be noted that the survival-related IRGs included in the IRGPI are referred to as hub IRGs.
Assessment of IRGPI and genetic alteration analysis
Patients were divided into high- and low-risk groups, depending on whether their IRGPI values are greater or less than the median value, respectively. The corresponding Kaplan-Meier (K-M) survival curves were constructed to demonstrate the overall survival (OS) within different risk groups. The ROC curve was also established in order to assess the sensitivity and specificity of the model. Additionally, univariate and multivariate analyses of survival were carried out for both IRGPI and clinicopathologic factors. These analyses were performed using the R package “survival” (19). We also explored the correlation between the expression of hub IRGs and the clinicopathologic factors. Moreover, genetic alteration analysis of hub IRGs was conducted on cBioPortal (https://www.cbioportal.org/) (24).
Evaluation of immune cell infiltration in tumors
The Tumor Immune Estimation Resource (TIMER) was used to systematically analyze immune cell infiltration in 23 types of cancer listed in the TCGA (25). Statistical methods were adopted to estimate the abundances of six immune cell infiltrates, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells. The obtained results were validated by pathological estimations. Subsequently, the immune cell infiltration levels in COAD samples were downloaded, and the correlation between IRGPI and immune cell infiltration was calculated based on Pearson correlation analysis.
Construction of the transcription factor (TF)-mediated regulatory network
TFs play an essential role in controlling gene expressions, including IRG, and thus, they can affect the immune function of the body. Consequently, it is necessary to identify the particular TFs that can potentially regulate hub IRGs. For this purpose, the list of TFs was first downloaded from the Cistrome Cancer database (http://cistrome.org/CistromeCancer/) (26). Then, the differentially expressed TFs were extracted from all DEGs. Subsequently, we carried out the correlation analysis of differentially expressed TFs and hub IRGs using the R software. The correlativity was considered reliable if |correlation value| >0.6 and P<0.05. As a result, we submitted them to build a TF-mediated regulatory network via Cytoscape software. To screen for the most important TF in this network, topological analysis was performed using the Network Analyzer plug-in (27).
Statistical analysis
To evaluate IRGPI effectiveness, the R package “survival” was used to calculate the area under curve (AUC) of the receiver operating characteristic (ROC) curve (28), and the R package “ggplot2” was used to generate boxplots (18). An independent t-test was conducted to test the differences among diverse clinical parameters. These differences were considered to be statistically significant if P<0.05.
Results
Acquisition of differentially expressed IRGs
In order to identify the DEGs, the genomic data of 473 COAD and 41 adjacent normal samples were compared using R package “limma”. Based on the cut-off criteria of |log2(fold change)| >1 and FDR <0.05, a total of 6,477 DEGs were detected, including 4,561 up-regulated genes and 1,916 down-regulated genes (Figure 1A,B). With the list of IRGs, 467 differentially expressed IRGs were extracted from all DEGs, including 179 up-regulated IRGs and 288 down-regulated IRGs (Figure 1C,D).
Functional enrichment analysis of differentially expressed IRGs
To explore the biological functions of differentially expressed IRGs, functional enrichment analysis was conducted using the DAVID 6.8. The results illustrated in Figure 2A show that “immune response”, “extracellular region”, and “antigen binding” are the most relevant BP, CC, and MF of genes, respectively. Furthermore, the KEGG enrichment analysis presented in Figure 2B indicates that the differentially expressed IRGs are mostly correlated with “cytokine-cytokine receptor interaction”.
Functional enrichment analysis of survival-related IRGs
As key prognostic indicators in applications of clinical tumor surveillance, molecular biomarkers related to IRGs could be used to evaluate the therapeutic efficiency of cancer immunotherapy. Based on univariate Cox analysis, 51 survival-related IRGs were distinguished (P<0.05). As shown in Figure 3A, the most relevant BP, CC, and MF terms of these genes are “regulation of response to stimulus”, “extracellular space”, and “receptor ligand activity”, respectively. Again, “cytokine-cytokine receptor interaction” was found to be the most significant signaling pathway (Figure 3B).
Prognostic values of survival-related IRGs
In order to better define the characteristics of survival-related IRGs, the prognostic values of these genes were calculated. The resulting forest plot presented in Figure 4 demonstrates that 40 genes are characterized by HR >1, whereas 11 genes have HR <1. This suggests that most survival-related IRGs were risk factors affecting the prognosis of COAD.
Development of IRGPI
In view of the results of the multivariate Cox regression analysis, the IRGPI was developed. This index was based on the following formula: risk score = [expression level of CD1B × (–2.09883)] + [expression level of SLC10A2 × 0.718209] + [expression level of FGF2 × 0.354982] + [expression level of CCL28 × (–0.1162)] + [expression level of IGHV4-31 × 0.00944] + [expression level of IGKV1-6 × 0.006782] + [expression level of ESM1 × 0.199876] + [expression level of TNFSF12 × 0.09248] + [expression level of UCN × 0.376204] + [expression level of VIP × 0.066404] + [expression level of GLP2R × (–4.08525)] + [expression level of IL1RL2 × 0.156916]. Depending on the scores and on the survival time, the patients were divided into high- (IRGPI > median value) and low-risk (IRGPI < median value) groups. As shown in Figure 5, the high-risk group exhibits more deaths than the low-risk one.
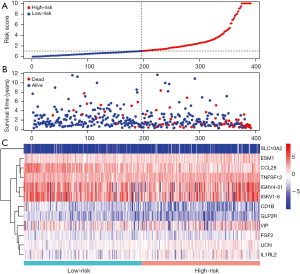
Evaluation of IRGPI
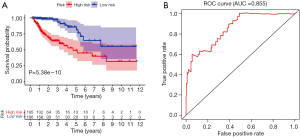
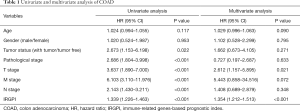
The calculated IRGPI values were used to determine the survival probability of COAD patients. As shown in Figure 6A, the OS of high-risk group patients was found to be less than that of the low-risk group patients (P<0.001). To assess the predictive accuracy of the IRGPI established herein, the ROC curve was generated, and the AUC was calculated (Figure 6B). This area was found to be 0.855, which indicates that the proposed IRGPI has excellent potential in monitoring prognosis. Moreover, the results of both univariate and multivariate analyses (Table 1) demonstrate that IRGPI is significantly correlated with survival, and thus, it can be used as an independent indicator when multiple clinicopathologic factors are taken into account.
Full table
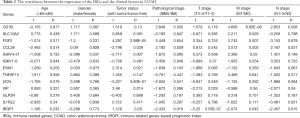
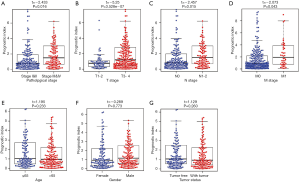
To further assess the clinical value of IRGPI, the relationships between the hub IRGs implicated in IRGPI and the clinicopathologic factors were analyzed. As shown in the results of Table 2, tumor status and M stage involved the expression of more genes. Besides, as an independent indicator, IRGPI had statistically significant difference in pathological stage (Figure 7A), T stage (Figure 7B), N stage (Figure 7C), and M stage (Figure 7D), which suggested that IRGPI could make an accurate prediction in different pathological stages of COAD tumors. However, IRGPI showed no significant difference in age (Figure 7E), gender (Figure 7F) and tumor status (Figure 7G).
Full table
Mutation landscape of hub IRGs included in IRGPI
Considering the significant clinical value of hub IRGs, their molecular characteristics were comprehensively explored. Specifically, genetic alteration analysis of these genes was carried out using the cBioPortal. The obtained results indicate that the most frequent type of genetic alteration is the elevated expression of mRNA (Figure 8A). For most IRGs (75%), the genetic alteration rate was found to be ≥5%, with the ESM1 and IL1RL2 genes showing the highest alteration frequencies (Figure 8B).

Clinical application of IRGPI
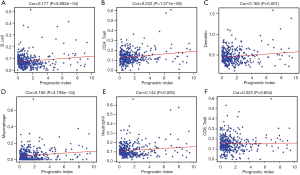
In order to estimate whether the IRGPI could accurately reflect the status of tumor immune microenvironments, we analyzed the relationship between IRGPI and the abundance of immune cell infiltration. The obtained results demonstrate that IRGPI is significantly correlated with 5 types of immune cells, including B cells (Figure 9A), CD4 T cells (Figure 9B), dendritic cells (Figure 9C), macrophages (Figure 9D), and neutrophils (Figure 9E). Moreover, there was no significant correlation between IRGPI and CD8 T cells (Figure 9F).
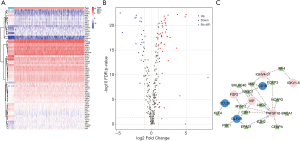
TF-mediated regulatory network
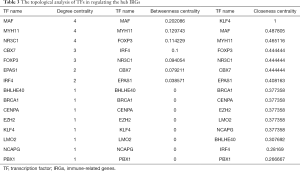
In order to explore the molecular mechanisms of hub IRGs, we studied their regulatory mechanism. Knowing that TFs play a key role in regulating the molecular network, the expression profiles of 318 TFs were examined, and 68 differentially expressed TFs were identified (Figure 10A,B). As a result, a TF-mediated regulatory network was constructed based on 15 TFs and 8 hub IRGs (Figure 10C). Afterwards, we performed a topological analysis on these TFs based on three centrality algorithms, including degree centrality, betweenness centrality and closeness centrality. As shown in Table 3, MAF, MYH11, and NR3C1 ranked the top 5 TFs in all three centrality analyses, and thus, they were considered as key TFs in the TF-mediated regulatory network.
Full table
Discussion
The development of targeted drugs and cancer immunotherapy has shifted COAD treatment from surgery and chemoradiotherapy to precise and individualized therapies (29). However, due to the limitations of immunotherapy treatments, it is necessary to identify the effective population and predict the clinical outcome beforehand. This may be accomplished by analyzing the immune genes and prognostic status of patients. In this study, we established an IRGPI that may be used to evaluate immune response in COAD patients, and thus, serves as a new biomarker for cancer immunotherapy.
Our results demonstrate that the most relevant BP of differentially expressed IRGs is “immune response”, and that the survival-related IRGs are highly associated with “the regulation of response to stimulus”. Based on KEGG enrichment analysis, the survival-related IRGs were significantly correlated with “cytokine-cytokine receptor interaction”, which suggested that the response of the immune system to stimuli is related to the expression of IRGs and patients’ survival. Furthermore, assessments of the prognostic efficiency of survival-related IRGs showed that most genes are characterized by high HR values, and thus, they affect the prognosis of COAD patients. This explains the variations in the efficacy of the same treatment between different patients.
Currently, the methods used to identify potential mechanisms and prognostic biomarkers are based on different types of bioinformatic analyses. For example, Dalerba et al. used the bioinformatics approach in combination with clinical-grade diagnostic assays to show that a subgroup of patients benefits from adjuvant chemotherapy (30). Other researchers used gene expression signatures based on mRNA, miRNA, or lncRNA expression profiles to predict the prognosis of COAD (31-33). In this study, we propose a biomarker based on IRGs, which is the most suitable way to reflect the immune status and tumor prognosis in patients. To establish this biomarker, the expression levels of survival-related IRGs were integrated using Cox regression analysis. Ultimately, 12 of 51 extracted genes were used to construct the predictive model that was used to calculate the risk scores of COAD patients. Based on these scores, the patients in the analyzed sample were divided into high- and low-risk groups.
The model was also used to establish an IRGPI. The reliability and efficiency of this index in terms of survival prediction was confirmed by ROC curve analysis. Univariate and multivariate analyses further prove the significant correlation between IRGPI and survival. Although the univariate analysis yields 5 independent indicators, only one (IRGPI) could be used in multivariate analysis. To test the feasibility of the IRGPI, the relationship between hub IRGs and multiple clinicopathological factors was also analyzed. The results indicate that as a comprehensive index, IRGPI exhibits stronger correlation with multiple pathological stages than single genes. Furthermore, the index is capable of accurate survival prediction in the pathological, T, N, and M stages.
Considering that cancer and immunity are closely associated, clinical treatments must take tumor immune microenvironments into account. Assessments of the correlation between the IRGPI and immune cell infiltration can be used to reflect the status of such microenvironments. Herein, we show that the IRGPI is significantly correlated with the infiltration levels of B cells, CD4+ T cells, dendritic cells, macrophages, and neutrophils (P<0.05). These immune cells play a very important role in COAD prognosis. For example, Gu et al. have recently shown that the elimination of pathogenic CD4+ T-cells and the induction of antitumor CD8+ T-cell activity can suppress colon carcinogenesis (34). Also, Vo et al. demonstrate that dendritic cell and lenalidomide combination therapy can effectively enhance antitumor immunity in a mouse COAD model (35). Moreover, according to Vinnakota et al., M2-like macrophages induce COAD cell invasion via matrix metalloproteinases (36).
To explore the molecular mechanisms underlying hub IRGs activity, a TF-mediated regulatory network was established. This network is comprised of several TFs that can simultaneously regulate the expressions of multiple IRGs, such as MAF, MYH11, and NR3C1. The available research indicates that c-MAF is capable of controlling immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4+ T cells (37). As for MYH11, it exhibits significantly different expressions in tumor samples, compared to normal tissues, and thus, it is considered as a potential tumor biomarker (38). Finally, the NR3C1 gene expressed during CD8+ T cell differentiation regulates the formation of memory-precursor cell fates (39). Other studies have also investigated the role of TFs in COAD prognosis. For instance, Wei et al. established a regulatory network of TF-miRNA-target genes to analyze disease progression and optimize treatment strategies (40). However, Mullany et al. showed that TF expression and related miRNAs influence the survival of patients diagnosed with colorectal cancer (41). Based on these studies and on our own results, we believe that TFs may play an important role in regulating the IRG expressions in COAD patients. Further investigation is needed to confirm this hypothesis.
Considering that the IRGPI established herein may be applied to accurately assess the immune status and monitor COAD prognosis in patients, it is actually an outstanding biomarker of the disease. This robust biomarker can be used in clinical prognosis applications, including cancer immunotherapy.
Acknowledgments
The authors would like to thank the TCGA, ImmPort, cBioPortal, and Cistrome cancer databases for the availability of the data needed for analyses.
Funding: This study was supported by grants from the Major Program of the National Natural Science Foundation of China (No. 81891012) and the National natural science foundation of China (No. U1812403-5-3).
Footnote
Conflict of Interests: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Banerjee A, Pathak S, Subramanium VD, et al. Strategies for targeted drug delivery in treatment of colon cancer: current trends and future perspectives. Drug Discov Today 2017;22:1224-32. [Crossref] [PubMed]
- Bonetti A, Giuliani J, Muggia F. Targeted agents and oxaliplatin-containing regimens for the treatment of colon cancer. Anticancer Res 2014;34:423-34. [PubMed]
- Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013;14:1014-22. [Crossref] [PubMed]
- Chen YP, Zhang Y, Lv JW, et al. Genomic analysis of tumor microenvironment immune types across 14 solid cancer types: immunotherapeutic implications. Theranostics 2017;7:3585-94. [Crossref] [PubMed]
- Sokratous G, Polyzoidis S, Ashkan K. Immune infiltration of tumor microenvironment following immunotherapy for glioblastoma multiforme. Hum Vaccin Immunother 2017;13:2575-82. [Crossref] [PubMed]
- Locy H, de Mey S, de Mey W, et al. Immunomodulation of the tumor microenvironment: turn foe into friend. Front Immunol 2018;9:2909. [Crossref] [PubMed]
- Newick K, O'Brien S, Moon E, et al. CAR T cell therapy for solid tumors. Annu Rev Med 2017;68:139-52. [Crossref] [PubMed]
- Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov 2019;18:197-218. [Crossref] [PubMed]
- Makkouk A, Weiner GJ. Cancer immunotherapy and breaking immune tolerance: New approaches to an old challenge. Cancer Res 2015;75:5-10. [Crossref] [PubMed]
- Roychowdhury S, Chinnaiyan AM. Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J Clin 2016;66:75-88. [Crossref] [PubMed]
- Li T, Kung HJ, Mack PC, et al. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol 2013;31:1039-49. [Crossref] [PubMed]
- Wang Z, Jensen MA, Zenklusen JC. A practical guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol 2016;1418:111-41. [Crossref] [PubMed]
- Jensen MA, Ferretti V, Grossman RL, et al. The NCI Genomic Data Commons as an engine for precision medicine. Blood 2017;130:453-9. [Crossref] [PubMed]
- Bhattacharya S, Andorf S, Gomes L, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res 2014;58:234-9. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847-9. [Crossref] [PubMed]
- Ito K, Murphy D. Application of ggplot2 to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol 2013;2:e79. [Crossref] [PubMed]
- Rizvi AA, Karaesmen E, Morgan M, et al. gwasurvivr: an R package for genome-wide survival analysis. Bioinformatics 2019;35:1968-70. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Vesztrocy AW, Dessimoz C. A gene ontology tutorial in python. Methods Mol Biol 2017;1446:221-9. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Zhang Z, Reinikainen J, Adeleke KA, et al. Time-varying covariates and coefficients in Cox regression models. Ann Transl Med 2018;6:121. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Tang Q, Chen Y, Meyer C, et al. A comprehensive view of nuclear receptor cancer cistromes. Cancer Res 2011;71:6940-7. [Crossref] [PubMed]
- Assenov Y, Ramírez F, Schelhorn SE., et al. Computing topological parameters of biological networks. Bioinformatics 2008;24:282-4. [Crossref] [PubMed]
- Kamarudin AN, Cox T, Kolamunnage-Dona R. Time-dependent ROC curve analysis in medical research: current methods and applications. BMC Med Res Methodol 2017;17:53. [Crossref] [PubMed]
- Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol 2015;33:1787-96. [Crossref] [PubMed]
- Dalerba P, Sahoo D, Paik S, et al. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N Engl J Med 2016;374:211-22. [Crossref] [PubMed]
- Sun D, Chen J, Liu L, et al. Establishment of a 12-gene expression signature to predict colon cancer prognosis. PeerJ 2018;6:e4942. [Crossref] [PubMed]
- Jacob H, Stanisavljevic L, Storli KE, et al. Identification of a sixteen-microRNA signature as prognostic biomarker for stage II and III colon cancer. Oncotarget 2017;8:87837-47. [Crossref] [PubMed]
- Xing Y, Zhao Z, Zhu Y, et al. Comprehensive analysis of differential expression profiles of mRNAs and lncRNAs and identification of a 14-lncRNA prognostic signature for patients with colon adenocarcinoma. Oncol Rep 2018;39:2365-75. [PubMed]
- Gu T, De Jesus M, Gallagher HC, et al. Oral IL-10 suppresses colon carcinogenesis via elimination of pathogenicCD4+ T-cells and induction of antitumor CD8+ T-cell activity. Oncoimmunology 2017;6:e1319027. [Crossref] [PubMed]
- Vo MC, Nguyen-Pham TN, Lee HJ, et al. Combination therapy with dendritic cells and lenalidomide is an effective approach to enhance antitumor immunity in a mouse colon cancer model. Oncotarget 2017;8:27252-62. [Crossref] [PubMed]
- Vinnakota K, Zhang Y, Selvanesan BC, et al. M2-like macrophages induce colon cancer cell invasion via matrix metalloproteinases. J Cell Physiol 2017;232:3468-80. [Crossref] [PubMed]
- Gabryšová L, Alvarez-Martinez M, Luisier R, et al. C-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4+ T cells article. Nat Immunol 2018;19:497-507. [Crossref] [PubMed]
- Lee WS, Seo G, Shin HJ, et al. Identification of differentially expressed genes in microsatellite stable HNPCC and sporadic colon cancer. J Surg Res 2008;144:29-35. [Crossref] [PubMed]
- Yu B, Zhang K, Milner JJ, et al. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat Immunol 2017;18:573-82. [Crossref] [PubMed]
- Wei S, Chen J, Huang Y, et al. Identification of hub genes and construction of transcriptional regulatory network for the progression of colon adenocarcinoma hub genes and TF regulatory network of colon adenocarcinoma. J Cell Physiol 2020;235:2037-48. [Crossref] [PubMed]
- Mullany LE, Herrick JS, Wolff RK, et al. Transcription factor-microRNA associations and their impact on colorectal cancer survival. Mol Carcinog 2017;56:2512-26. [Crossref] [PubMed]


