Contribution of m6A subtype classification on heterogeneity of sepsis
Introduction
Sepsis is defined as a highly heterogeneous syndrome that is associated with a dysregulated systemic inflammatory host response to infection and causes organs dysfunction (1,2). It is generally believed that the failure of proposed sepsis therapies is due to the heterogeneity of sepsis patients and the inability to accurately classify sepsis patients at the molecular biology level. The identification of distinct subtypes of sepsis could adopt appropriate treatment regimens for patients with different subtypes, which can improve survival in sepsis (3,4).
Previous studies attempted to identify diverse subtypes through clinical features or biomarkers. For example, Seymour et al. classified sepsis patients to four derived phenotypes via 29 clinical features (temperature, mean arterial pressure, fluid resuscitation response and central venous oxygen saturation, etc.). Using different genes between sepsis and health controls, researchers identified four subphenotypes of sepsis, among them. However, these classification studies of sepsis have not uncovered the mechanism of different subtypes. N6-methyladenosine (m6A), a chemical modification for RNA regulation, accounts for more than 60% of all RNA epigenetics (5,6). As m6A is the commonest posttranscriptional RNA modification involved in the regulation of whole bioprocesses (including immunity, metabolism, proliferation and apoptosis), the heterogeneity in sepsis may associate with m6A RNA regulations. In addition, since m6A emerged as a reversible bioprocess involved in many various disease progressions, potential therapeutic targets of m6A were widely investigated (7-11). Previous studies have demonstrated that m6A regulators were closely associated with tumor heterogeneity (12,13). However, details of m6A RNA regulatory factors mediating sepsis heterogeneity remained largely unknown. Fortunately, the public database provides a large number of sepsis-related transcriptome data, while the use of RNA omics and RNA-seq are becoming cheaper and more extensive, helping us to perform integrative analysis on m6A RNA methylation and sepsis molecules subtypes.
In this scenario, we systematically analyzed the expression of widely reported m6A RNA methylation regulators in sepsis based on public database- Gene Expression Omnibus (GEO database) (5-16). Firstly, we sought to screen prognostic m6A RNA methylation regulators in sepsis through univariate Cox’s proportional-hazards regression. Furthermore, we classified sepsis into diverse m6A endotypes through unsupervised clustering according to prognostic m6A molecules. In addition, we attempted to elaborate the reasons of different prognosis in m6A induced subtypes via enrichment analysis.
Methods
Data sources and study selection
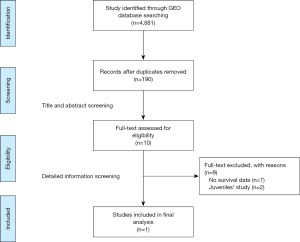
The public database—GEO database was searched for all expression microarrays that matched terms of sepsis. The datasets collected from clinical studies investigating sepsis in adults using peripheral blood within 48 hours after intensive care unit admission (ICU) were included. Exclusion criteria were as follows: (I) datasets that utilized endotoxin or lipopolysaccharide infusion as vitro or animal models for sepsis; (II) clinical-gene expression microarray derived from sorted cells; (III) without the outcome of 28 day mortality or survival curves. The flow-process diagrams for screening datasets were shown in Figure S1.
Collecting m6A RNA methylation regulators via systematic review
A systematic literature review of all of the pertinent English language studies was undertaken in the PubMed databases from inception through December 28, 2019. A manual search of the selected articles and relevant review articles was performed by two reviews (F Liu and ZS Wu) independently. Only m6A RNA methylation regulators gene which confirmed by animal or cell experiments will eligible to present analysis.
Data preprocessing
All datasets were downloaded as txt files, and outputs from mRNA array were normal-exponential background corrected and then between-arrays quantile normalized using limma R package. For compatibility with microarray study, expression was normalized using a weighted linear regression, and the estimated precision weights of each observation were then multiplied with the corresponding log2 to yield final gene expression values.
Screening prognosis-associated m6A RNA methylation regulators in sepsis
In order to determine prognostic m6A RNA methylation regulators, the univariate Cox’s proportional-hazards regression with Bonferroni correction for multiple comparisons was utilized, using survival R package, with cut-off value as P<0.05.
Explore m6A induced molecular subtypes of sepsis
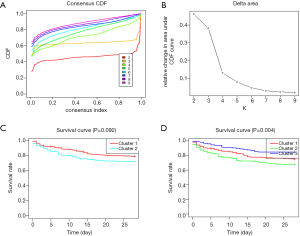
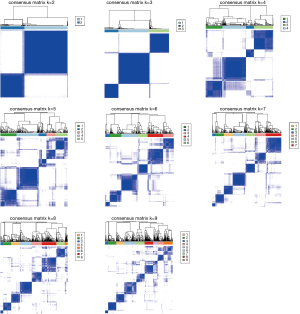
The consensus k means clustering was utilized to perform consistent clustering and selecting of m6A subtypes based on prognostic m6A RNA methylation regulators expression profiles. The clustering was performed using 100 iterations, with each iteration containing 80% of samples. The optimal cluster number was determined by cumulative distribution function (CDF) curves of the consensus score, clear separation of the consensus matrix heatmaps, characteristics of the consensus cumulative distribution function plots, and adequate pair wise–consensus values between cluster members.
Assessment for immune heterogeneity among subtypes
Kaplan-Meier (KM) curves would be performed to evaluate prognosis of various m6A Subtypes, with cut-off value as P<0.05. Differential expression analysis using moderated t-tests would be utilized to assess the expression distribution of twelve m6A RNA methylation regulators in different subtypes.
We furthermore evaluated the variation of immune status among subtypes. Gene Set Enrichment Analysis (GSEA) and cell type identification by estimating relative subset of known RNA transcripts (CIBERSORT) were utilized to evaluate proportions of immune cells. Briefly, GSEA calculates separate enrichment scores (ES) for each pairing of a sample and gene set (17) via GSVA and GSEABase R package. Each GSEA ES represents the degree to which the genes in a particular gene set are coordinately up- or downregulated within a sample. CIBERSORT algorithm could calculate proportions of human immune cells according to RNA matrix (https://cibersort.stanford.edu/) (18). Immune cells included immune enhancing cells (Th1 cells, T cells CD4 activated, NK cells activated and B cells activated) and immune suppressive cells (Th2 cells and Treg cells). Differential expression analysis using moderated t-tests would be utilized to assess the expression distribution of pro-inflammatory cytokines and enrichment of immune cells among subgroups, P<0.05 was recognized as significant results. Pro-inflammatory cytokines included interleukin-1β (IL-1β), interleukin-6 (IL-6) and Tumor necrosis factor (TNF).
Assessing the heterogeneity of biological function among subtypes
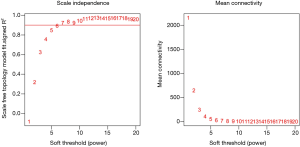
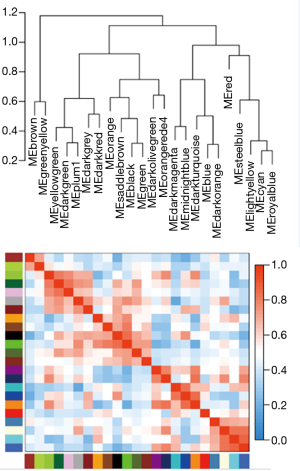
There are two steps to implement this analysis. On the one hand, we attempted to find gene-sets which significantly correlated to m6A subtypes through Weighted gene co-expression network analysis (WGCNA), using WGCNA R package to determine co-expressed genes using all expressed genes in microarrays (19,20). Analysis setting included bi-weight miscorrelation (corType = ‘bicor’) to account for outliers, sign of correlations between neighbors (TOM type and networkType = ‘signed’), and a more sensitive module detection parameter (deepSplit = 3). Module eigengene (ME) was calculated as the first principal component of gene expression for the module and inter-relatedness of each module by eigengene network clustering (Figure S2). MEs were compared with m6A subtype information using Spearman’s correlation corrected for subtypes, and P values were adjusted for multiple comparisons by false discovery rate. Genes from modules which highly associated with m6A subtypes (the maximum correlation coefficient and P<0.05) were selected for further Gene Ontology (GO) analysis.
On the other, GO analysis was performed to elaborate the functions of selected gene-sets using org.Hs.eg.db, clusterProfiler and enrichplot R packages, with cut-off value as P<0.05.
Co-expression analysis was performed to furthermore explore the possible downstream molecules regulated by m6A RNA methylation, with cut-off value as P<0.05, Pearson correlation coefficient >0.6.
Software and versions
Rx64 3.6.1 was conducted to process data, analyze data and plot diagrams; Cytoscape 3.6.1 was performed to plot network diagrams.
Results
Characteristics of datasets and patients
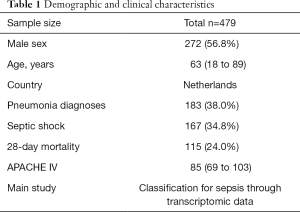
After search strategy and inclusive criteria, one mRNA dataset from University Medical Center in Utrecht and Academic Medical Center in Amsterdam is finally enrolled in current study. A total of 479 patients in this dataset were all older than 18 years and were diagnosed with sepsis. Septic shock ratio is 34.8%, and more details are shown in Table 1.
Full table
Twelve m6A RNA methylation regulators were collected via systematic review
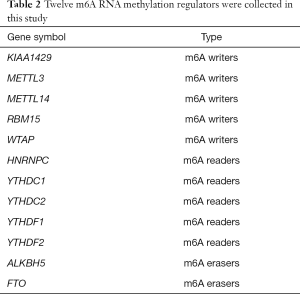
We first collated a list of sixteen m6A RNA methylation regulators from published literature (5-16), and then we restricted the list to genes with available RNA expression data in the GEO datasets. This yielded a total of 12 m6A RNA methylation regulators. These m6A RNA regulators included m6A writers, m6A readers and m6A erasers. m6A writers included KIAA1429, methyltransferase like 3 (METTL3), methyltransferase like 14 (METTL14), RNA binding motif protein 15 (RBM15) and WT1 associated protein (WTAP). m6A readers included HNRNPC, YTH domain containing 1 (YTHDC1), YTH domain containing 2 (YTHDC2), YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) and YTH N6-methyladenosine RNA binding protein 2 (YTHDF2). m6A erasers included alkB homolog 5, RNA demethylase (ALKBH5) and FTO alpha-ketoglutarate dependent dioxygenase (FTO), shown in Table 2.
Full table
The prognosis associated m6A RNA methylation regulators in sepsis
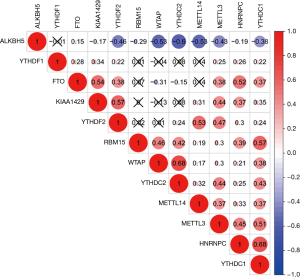
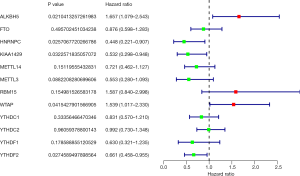
According to the univariate Cox regression analyses, ALKBH5, HNRNPC, KIAA1429, WTAP and YTHDF2 are significantly correlated with 28-day cumulative mortality (P<0.05). ALKBH5 and WTAP are risky genes with Hazard Ratio (HR) >1, and HNRNPC, KIAA1429 and YTHDF2 are protective genes with HR <1 (Figure 1). The correlation results among m6A RNA methylation regulators were shown in Figure S2.
m6A molecular subtypes in sepsis
Based on the expression similarity of m6A RNA methylation regulators, k =2 or 3 seemed to be an adequate selection with clustering stability increasing from k =2 to 9 in cohort (Figure 2 and Figure S3). KM suggested significantly different 28-day cumulative mortality among three subtypes (P=0.004). However, 28-day cumulative mortality did not show an obviously statistic difference between two subtypes (P=0.092). In consideration of KM curves and clustering stability, patients with sepsis were finally divided into three subtypes (cluster 1, cluster 2 and cluster 3).
In the sepsis cohort, the best outcome was found for patients classified as having a cluster 3, and at 28 days, 21 (14.6%) of 144 people with a cluster 3 had died (HR vs. all other clusters 5.42, 95% CI: 0.359–0.819; P=0.011), compared with 57 (25%) of 224 people with a cluster 1 (HR 0.579, 95% CI: 0.364–0.920; P=0.037), and 36 (23%) of 112 people with a cluster 2 (HR 0.477, 95% CI: 0.272–0.833; P=0.003) .
Immune status heterogeneity among m6A subtypes
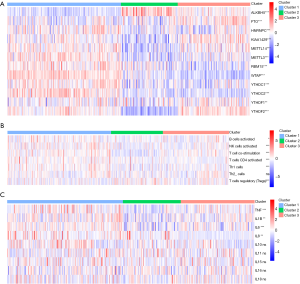
Figure 3A showed a distinct expression pattern in the m6A regulators profiles of each subtype (P<0.05). After GSEA and CIBERSORT, the obviously heterogeneity of immune status among subtypes were identified in Figure 3B,C.
This heterogeneity of immune status included significant difference of enrichment scores in immune cells (Figure 3B), and obvious discrepancy of expression in pro-inflammatory cytokines (Figure 3C). In cluster 1 (moderate prognosis), immune enhancing cells and pro-inflammatory cytokines were significantly up-regulated compared with other subtypes, companied with immune suppressive cell down-regulated simultaneously (P<0.05). The distribution of immune cells and pro-inflammatory cytokines in cluster 2 (the worst prognosis) showed diametrically opposed to those in cluster 1 (P<0.05). The enrichment score of immune cells and expression of pro-inflammatory cytokines in cluster 3 (the best prognosis) were in the middle of cluster 1 and cluster 2.
Heterogeneity of other biological process among subtypes
WGCNA identified 24 modules in the sepsis cohort (Figures S4,S5). Cluster 1 positively correlated with Seventeen module (including 1,860 genes), with correlation coefficient as 0.73 and P value as 8×10−81. Cluster 2 markedly positively associated with One module (including 3,423 genes), with correlation coefficient as 0.68 and P value as 8×10−70. Cluster-3 significantly positively related to Twenty-one (including 251 genes), with correlation coefficient as 0.61 and P value as 8×10−51. Genes from these strongly relevant modules defined as the eigengenes of each subtypes (Figure 4A).
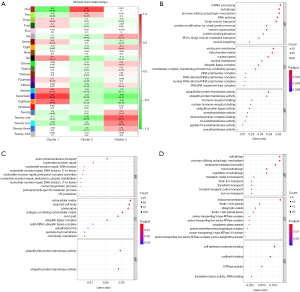
In order to furthermore explore module biological function, GO enrichment analyses were conducted and the results were shown in Figure 4B,C,D. The biological functions of Seventeen module (correlated with cluster 1) were mainly enriched in RNA processing and vesicle transport; One module (correlated with cluster 2) were mainly related to nucleotide repair and extracellular matrix; Twenty-one (correlated with cluster 3) were mainly associated with autophagy.
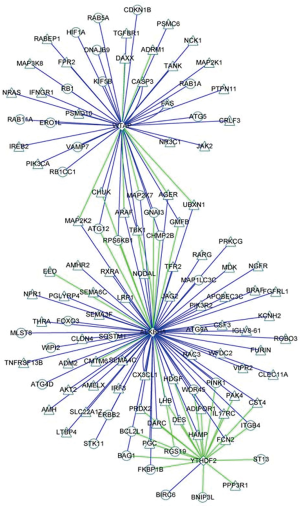
Co-expression analysis furthermore identified 82 immune molecules and 40 autophagy-related molecules could be regulated by prognostic m6A RNA methylation regulators, P<0.05 and correlation coefficient >0.6 (Figure S6). The immune molecules and autophagy-related molecules were referenced to immunology database (https://www.immport.org/) and human autophagy database (http://www.autophagy.lu/), respectively.
Discussion
Sepsis is a clinically common syndrome with high heterogeneity. Heterogeneity in patients with sepsis is currently considered to be an important cause of treatment failure. Therefore, it is very important to find a regulatory analysis that affects the heterogeneity of concentration and then guide the treatment of sepsis. Current research indicates that the expression of RNA methylation, regulators of epigenetic, is closely related to the heterogeneity and prognosis of sepsis. Three m6A molecular subtypes were identified in sepsis, cluster 1/2/3, by consensus clustering based on the expression of most aberrant m6A regulators. Three m6A subtypes exhibited significantly different RNA epigenetics, immune status, biological processes and outcomes. We further investigated that the moderate immune activated status and potential autophagy mechanisms could benefit septic patients, which were regulated by RNA methylation. To our knowledge, this was the first study on the transcriptome-wide mapping of m6A regulators which focuses on investigating the landscapes and functions of the reversible RNA modifications in sepsis.
YTHDF2, HNRNPC, KIAA1429, WTAP, and ALKBH5 were proved to be more important in prognosis of sepsis. The major mechanism by which m6A exerts its effects is by recruiting m6A-binding proteins (m6A readers) to target RNAs. Subsequently, a methyltransferase complex within the nuclear speckle, m6A writers, installs m6A modification on targeted RNAs via the methyl groups of S-adenosylmethionine (SAM) transferase. Demethylases (erasers) were found that removes methyl groups from m6A, indicating that m6A is a dynamically reversible RNA modification. The previous studies identified YTHDF2 and HNRNPC as m6A readers, KIAA1429 and WTAP as m6A writers and ALKBH5 as m6A eraser (4-14). The current study further uncovered the prognostic roles of these five m6A regulators in sepsis. The future studies could be conducted to sequentially verify and research these m6A elaborate mechanisms in sepsis.
The current study provided an insight that immune heterogeneity of sepsis was largely associated with m6A RNA methylation. Patients with same syndrome (sepsis) were classified into finer taxa and had differential prognosis and distinct immune status. Patients in cluster 2 with worst prognosis suffered from immunosuppression; patients in cluster 1 with worse prognosis suffered from hyper-activated immunocompetent status; and patients in cluster 3 (the best prognosis) had the moderate immune activity. The extent of hyper-activated and hypo-activated immune response varies among individuals in sepsis, which result in immune heterogeneities (3,4). It is well known that hyper-activated immunocompetent and Immunosuppression are both detrimental status in the progression of sepsis (5,7), especially immunosuppression. However, for a given patient, it is too hard to characterize the immune status due to complex and unknown mechanisms of immune heterogeneity in sepsis. As immune functions were widely regulated by m6A RNA methylation (9,10), three m6A subtypes enhanced the comprehension of molecular characteristics and subgroup-specific immune status in highly heterogeneous syndrome (sepsis). These results provided a better understanding of the predicting clinical outcomes and valuable research targets in sepsis.
The molecular characterization of the transcriptome of subtypes subsequently indicated that autophagy mechanisms, regulated by RNA methylation, could play an important role in improvement of outcomes in sepsis, which were consistent with previous studies of autophagy in sepsis. Autophagy is the regulated process cells use to recycle nonessential, redundant, or inefficient components and is an adaptive response during times of stress (21). In addition to its function in enabling the cell to gain vital nutrients in times of stress, autophagy can also be involved in elimination of intracellular microorganisms, tumor suppression, and antigen presentation (22). Therefore, previous studies also confirmed that autophagy-related processes were properly activated and accelerated during sepsis due to the sepsis induced mitochondrial injury (22). Besides, autophagy was widely recognized as protective biological processes in sepsis (21-28). Unfortunately, these autophagy-related studies did not show upstream regulatory mechanisms. As autophagy seemed to be not reversible once triggered, exploration for regulatory factors of autophagy would be beneficial for develop potential therapeutic targets in sepsis. Combined with current analysis and previous study, outcomes of septic patients in cluster 3 were obviously improved largely depending on autophagy, which were quite possibly regulated by m6A RNA methylation.
As m6A RNA methylation is generally known a reversible and regulatable process, chemicals targeting m6A methylation would be explored as a new therapeutic method for sepsis therapy. The present study provided possible regulatory relationships between m6A regulators with immune and autophagy-related molecules.
There are several limitations in the present study. First, as a retrospective study of primarily publically available data, we were not able to download more demographics and clinical features such as severity, complications, individual treatment of each patient for detailed and longitudinal analyses. In addition, for the limitation of bioinformatics, the further studies and experiments should be conducted to sequentially verify and research elaborate RNA methylation mechanisms screened by our analyses.
Conclusions
We identified three highly heterogenous m6A subtypes in sepsis, with significantly different RNA epigenetics, immune status, biological processes and outcomes, which could be intervention targets for improvement of therapeutic system in sepsis, with validation experiments to be warranted to assess these further.
Acknowledgments
We would like to thank all of the microarray data contributors of this study and all of the patients and volunteers who participated in this study.
Funding: The study was supported in part by the National Natural Science Foundation of China (81571847, 81930058), the projects of Jiangsu Provincial Medical Key Discipline (ZDXKA2016025) and Jiangsu Provincial Special Program of Medical Science (BE2018743, BE2019749).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study was not required ethics approval since it is a secondary bioinformatics analysis based on public database (GEO database).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Reinhart K, Daniels R, Kissoon N, et al. Recognizing sepsis as a global health priority - A WHO resolution. N Engl J Med 2017;377:414-7. [Crossref] [PubMed]
- Fleischmann C, Scherag A, Adhikari NK, et al. Assessment of global incidence and mortality of hospital-treated sepsis: current estimates and limitations. Am J Respir Crit Care Med 2016;193:259-72. [Crossref] [PubMed]
- Davenport EE, Burnham KL, Radhakrishnan J, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med 2016;4:259-71. [Crossref] [PubMed]
- Seymour CW, Kennedy JN, Wang S, et al. Derivation, Validation, and Potential Treatment Implications of Novel Clinical Phenotypes for Sepsis. JAMA 2019;321:2003-17. [Crossref] [PubMed]
- Meyer KD, Jaffrey SR. Rethinking m(6)A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol 2017;33:319-42. [Crossref] [PubMed]
- Berlivet S, Scutenaire J, Deragon JM, et al. Readers of the m(6)A epitranscriptomic code. Biochim Biophys Acta Gene Regul Mech 2019;1862:329-42.
- Maity A, Das B. N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. FEBS J 2016;283:1607-30. [Crossref] [PubMed]
- Tuncel G, Kalkan R. Importance of m N(6)-methyladenosine (m(6)A) RNA modification in cancer. Med Oncol 2019;36:36. [Crossref] [PubMed]
- Zhang C, Fu J, Zhou Y. A Review in Research Progress Concerning m6A Methylation and Immunoregulation. Front Immunol 2019;10:922. [Crossref] [PubMed]
- Dang W, Xie Y, Cao P, et al. N(6)-Methyladenosine and Viral Infection. Front Microbiol 2019;10:417. [Crossref] [PubMed]
- Sweeney TE, Perumal TM, Henao R, et al. A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun 2018;9:694. [Crossref] [PubMed]
- Chai RC, Wu F, Wang QX, et al. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging (Albany NY) 2019;11:1204-25. [Crossref] [PubMed]
- Zhou J, Wang J, Hong B, et al. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma - a retrospective study using TCGA database. Aging (Albany NY) 2019;11:1633-47. [Crossref] [PubMed]
- Vu LP, Pickering BF, Cheng Y, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 2017;23:1369-76. [Crossref] [PubMed]
- Rosa-Mercado NA, Withers JB, Steitz JA, et al. Settling the m(6)A debate: methylation of mature mRNA is not dynamic but accelerates turnover. Genes Dev 2017;31:957-8. [Crossref] [PubMed]
- Li HB, Tong J, Zhu S, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017;548:338-42. [Crossref] [PubMed]
- Reimand J, Isserlin R, Voisin V, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 2019;14:482-517. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-457. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Zhang S, Wu Z, Xie J, et al. DNA methylation exploration for ARDS: a multi-omics and multi-microarray interrelated analysis. J Transl Med 2019;17:345. [Crossref] [PubMed]
- Watanabe E, Muenzer JT, Hawkins WG, et al. Sepsis induces extensive autophagic vacuolization in hepatocytes: a clinical and laboratory-based study. Lab Invest 2009;89:549-61. [Crossref] [PubMed]
- Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005;120:159-62. [PubMed]
- Chien WS, Chen YH, Chiang PC, et al. Suppression of autophagy in rat liver at late stage of polymicrobial sepsis. Shock 2011;35:506-11. [Crossref] [PubMed]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008;132:27-42. [Crossref] [PubMed]
- Schmid D, Munz C. Immune surveillance of intracellular pathogens via autophagy. Cell Death Differ 2005;12 Suppl 2:1519-27. [Crossref] [PubMed]
- Yan L, Vatner DE, Kim SJ, et al. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci USA 2005;102:13807-12. [Crossref] [PubMed]
- Chung KW, Kim KM, Choi YJ, et al. The critical role played by endotoxin-induced liver autophagy in the maintenance of lipid metabolism during sepsis. Autophagy 2017;13:1113-29. [Crossref] [PubMed]
- Qiu P, Liu Y, Zhang J. Review: the Role and Mechanisms of Macrophage Autophagy in Sepsis. Inflammation 2019;42:6-19. [Crossref] [PubMed]