
Dynamic analysis of N-glycomic and transcriptomic changes in the development of ovarian cancer cell line A2780 to its three cisplatin-resistant variants
Introduction
Ovarian cancer is the seventh most common cancer in women around the world and its mortality rate is second among gynecological cancers (1). Platinum and paclitaxel combination chemotherapy is a first-line postoperative regimen for ovarian cancer patients. Although 80% of patients have an initial response to platinum compounds, up to 70% of ovarian cancer patients will relapse and become resistant to platinum in the process of chemotherapy (2). Platinum resistance development is a dynamic process that occurs during continuous cycles of chemotherapy. Therefore, studies on the dynamic process involved in the development of platinum resistance should be performed during chemotherapy. This may provide a better understanding of platinum resistance, identify biomarkers needed to monitor platinum resistance, guide ovarian cancer patient treatment, and improve patient 5-year survival. For those ovarian cancer patients resistant to chemotherapy, monitoring is vital for avoiding delays in the effective treatment and unnecessary chemotherapy side effects (3). Thus, it is urgent to develop useful biomarkers for monitoring platinum resistance and to study the development of platinum resistance (4).
Glycosylation is one of the most common posttranslational modifications and is responsible for modulating and controlling the biological roles of glycoproteins and plays an important role in many physiological and pathological processes (5-8). Abnormal glycosylation related to drug resistance has been reported in several research studies (9-11). First, the sensitivity to platinum was altered in ovarian cancer after glycosylation of drug transporters was inhibited, such as P-glycoprotein, breast cancer resistance protein (BCRP, ABCG2), multidrug resistance protein 1 (MRP1, ABCC1), and multidrug resistance protein 4 (MRP4, ABCC4) (12,13). Second, aberrant glycosylation in some signaling pathways is also involved in the mechanisms of drug resistance. For instance, the phosphoinositide 3 kinase (PI3K)/Akt pathway was aberrantly activated by the glycosylation modification result in multidrug resistance (14,15). Third, few serum samples, provided by late-stage recurrent ovarian cancer patients before and after receiving an experimental drug treatment (i.e., a combination of docetaxel and imatinib mesylate), were analyzed to identify specific changes in protein glycosylation related to cancer progression. The results indicated that altered glycosylation of serum glycoprotein may be a potential biomarker for progression of ovarian cancer (16). Therefore, glycans are important predictive biomarkers and are crucial for the mechanism of drug resistance.
To the best of our knowledge, few glycomic analyses were performed on drug-resistant ovarian cancer and none of them focused on the dynamic process of chemotherapy (17). Due to heterogeneity and accessibility limitations of the clinical samples, a cancer cell line was chosen as an experimental model to study N-glycan alterations during continuous exposure to cisplatin. Moreover, cancer cells are more stable than clinical samples and are more convenient for mechanistic research (18). The present study mimics the development of platinum resistance development in ovarian cancer by continuously exposing A2780 cells to cisplatin. Cisplatin-resistant variants were established after 20, 30, and 40 continuous exposures to cisplatin and were confirmed by higher half maximal inhibitory concentration (IC50) values and increased ABCB1 expression compared to A2780 cells. Analysis of N-glycomic dynamic changes during the development of platinum resistance in cisplatin-resistant variants was performed using MALDI-time-of-flight (TOF)-MS. N-glycan alterations associated with platinum resistance were further validated on a glycotransferase level using transcriptome sequencing and real-time PCR (RT-PCR). To date, this is the first attempt to discover specific N-glycan changes in ovarian cancer cells during continuous exposure to cisplatin, which may provide more information about glycan biomarkers of platinum resistance and related cisplatin resistance mechanisms.
Methods
Cell culture and reagents
An ovarian cancer cell line A2780 and three cisplatin-resistant variants were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (BasalMedia Technologies, Shanghai, China) supplemented with 10% fetal bovine serum (Moregate Biotech, Melbourne, Australia) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, USA) at 37 °C in a humidified atmosphere of 5% CO2. Three cisplatin-resistant variants of A2780 cells were regularly stimulated with 3 µg/mL cisplatin (Abcam, Cambridge, UK) and subcultured once in drug-free medium before the experiment.
In this study, Milli-Q water (MQ) was generated from a Q-Gard 2 system (Millipore, Amsterdam, Netherlands). Sodium dodecyl sulfate (SDS), phosphate buffered saline (PBS), ethanol, and trifluoroacetic acid (TFA) were purchased from Merck (Merck, Darmstadt, Germany). Sodium hydroxide (NaOH), urea, dithiothreitol, ammonium bicarbonate, iodoacetamide, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimid (EDC) hydrochloride, and 1-hydroxybenzotriazole (HOBt) hydrate were obtained from Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO, USA). Recombinant peptide-N-glycosidase F (PNGase F), Nonidet P-40 (NP-40), and denaturation buffer were bought from Roche Diagnostics (Roche Diagnostics, Mannheim, Germany). Acetonitrile (ACN) and 2,5-dihydroxybenzoic acid (2,5-DHB) were from Bruker Daltonics (Bruker Daltonics, Bremen, Germany) and Biosolve (Biosolve, Valkenswaard, Netherlands), respectively.
Cell selection strategy
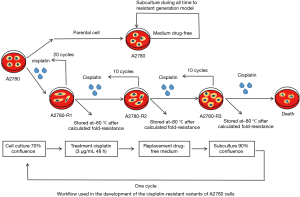
A2780 cells were plated in duplicate into six-well plates at a density of 5×105 cells per well and a control plate was set up separately. Once the cell culture reached 70% confluency, 2 mL of media with 3 µg/mL cisplatin were added to all plates excluding the control, which received 2 mL of drug-free medium. After incubation for 48 h at 37 °C with 5% CO2, the drug was removed and replaced by fresh drug-free culture medium. Once the cell culture reached 90% confluency, cells from one control plate and one drug plate were removed and counted. Cells from both plates were re-plated in duplicate into a six-well plate at a density of 5×105 cells per well and cultured until reaching 70% confluency for the next cisplatin exposure. These steps were repeated until reaching a total number of 40 cycles. The fold resistance were calculated when the 20th, 30th, and 40th cycles were done, which showed significant differences compared to A2780 cells. The three cisplatin-resistant variants were stored at –80 °C and named A2780-R1, A2780-R2, and A2780-R3, respectively. Cells were thawed together for cytotoxicity analysis and fold resistance calculations (19,20) (Figure 1).
CCK8 assay
Cell growth and viability were measured using the CCK8 assay (Dojindo, Kumamoto, Japan). Briefly, A2780 cells and three cisplatin-resistant variants were seeded into 96-well plates (5,000 cells/well). After 12 h, cells were treated with various concentrations of cisplatin (0, 0.5, 1, 2, 4, 8, 16, 32, and 64 µg/mL) for 48 h. Then, 10 µL of the CCK8 reagent were added to the cells and absorbance at 450 nm was measured by an EPOCH ELISA reader (BioTek Instruments, Vermont, USA) according to the manufacturer’s instructions.
Cell lysis preparation for mass spectrometry (MS) analysis
Cells were rinsed twice with PBS. After washing, 2% SDS-containing protease inhibitor cocktail (Roche Diagnostics, Roche Applied Science, Meylan, France) was used to lyse the cells on ice for 30 min. The lysate was then boiled for 15 min before centrifugation at 14,000 g for 30 min and the supernatant was collected for protein concentration quantification using a BCA kit (Thermo Scientific, San Jose, CA, USA).
Release and purification of N-glycans from glycoprotein
A total of 1 mg of protein from the supernatant was added to 200 µL of 2% SDS-containing 8-M urea and 10-mM dithiothreitol. The sample was heated to 56 °C for 20 min and then incubated in 40-mM ammonium bicarbonate containing 25-mM iodoacetamide at 37 °C for 30 min in the dark. Subsequently, the sample was transferred to an ultrafiltration unit (Amicon Ultra-0.5, Ultracel-10 membrane; Millipore, Billerica, MA, USA) and centrifuged at 14,000 g for 15 min. The sample was washed by adding 200 µL of 1× PBS to the ultrafiltration unit. Afterwards, 3–5 µL of PNGase F, 20 µL of NP-40, 20 µL of denaturation buffer, and 160 µL of 1× PBS were added to the device and incubated with 250 rpm shaking for 24 h at 37 °C. The ultrafiltration unit was transferred to a new collection tube before centrifuging at 14,000 g for 15 min. The filter membrane was then washed with 200 µL of 1× PBS three times. N-glycans from the collection tube were purified and desalted using Porous Graphic Carbon Solid-Phase Extraction (PGC-SPE). The PGC-SPE microcolumn, a GELoader tip filled with porous graphic carbon powder, was preconditioned with three volumes of 0.1% (v/v) TFA in 80% (ACN)/H2O (v/v) and equilibrated with three volumes of 0.1% (v/v) TFA. The sample was passed through the microcolumn five times to achieve complete glycan adsorption. Subsequently, the microcolumn was washed with two volumes of water to remove salts and buffer. Finally, N-glycans were eluted with 200 µL of 25% (v/v) ACN containing 0.05% (v/v) TFA and lyophilized in a vacuum freeze dryer (Martin Christ GmbH, Osterode, Germany).
Derivatization and enrichment of N-glycans
Lyophilized glycans were added to 100 µL of 250-mM EDC and 250-mM HOBt in ethanol and incubated for 1 h at 37 °C. Then, 100 µL of ACN were added and the mixture was incubated at –20 °C for 15 min. Subsequently, the mixture was brought to room temperature before glycan purification using a cotton HILIC microtip. For purification of the derived N-glycans, 10-µL pipette tips were packed with a 3-mm cotton thread. Cotton HILIC tips were pre-wetted three times with 10 µL of MQ water and then equilibrated with 10 µL of 85% ACN three times. Samples were loaded by carefully pipetting up and down 50 times. The tips were washed three times with 10 µL of 85% ACN/1% TFA, and three times with 10 µL of 85% ACN. In the end, N-glycans were eluted in 10 µL of MQ water by pipetting up and down 50 times (18,21).
MALDI-TOF-MS analysis
The TOFMixTM (LaserBio Laboratories, France) was used for calibration before MS analysis. A total of 5 µL of derivatized N-glycans were premixed with 5 µL of sDHB matrix (5 mg/mL in 50% ACN with 1-mM NaOH) and 2 µL of the mixture were spotted on a MALDI target plate (800/384 MTP AnchorChip, Bruker Daltonics, Bremen, Germany). Each sample was spotted in three replicates and spots were allowed to air dry at room temperature, followed by a MALDI-TOF-MS measurement. Profiles were generated by two laser shots and a total of 200 profiles were accumulated from different points of laser irradiation into one file for each sample spot.
MS data for N-glycans was pre-processed, normalized and extracted using the Progenesis MALDI software (Shimadzu Biotech, Kyoto, Japan). Relative intensity for each N-glycan was calculated by dividing the intensity of that N-glycan by the sum intensity of all identified N-glycans. Relative standard deviation (RSD) for relative intensity values was used to estimate the MS stability. In this study, RSD for the 77 N-glycans was <25%. The threshold of intensity ratios was set at 1.2 and 0.83 to estimate up- or down-regulation, respectively. Significant differences in N-glycans and glyco-subclasses were analyzed using Student’s t-test and P<0.05 was considered statistically significant. The proposed structures of identified glycans were interpreted manually based on their m/z values according to several published studies, GlycoWorkbench, and Glycan Mass Spectral Database.
Total RNA extraction and transcriptome sequencing
Total RNA was extracted using the TRIzol™ reagent (Invitrogen, Carlsbad, CA, USA). Transcriptome sequencing was conducted by OE Biotech Co., Ltd. (OE Biotech Co., Ltd, Shanghai, China). Glycogenes were searched and selected using the following keywords: “glyco”, “glycan”, “gluc”, “gal”, “mgat”, “st6”, “st3”, “gnt”, “galnt”, “fuc”, “mannose”, and “alg”.
Quantitative RT-PCR (qRT-PCR) analysis
Total RNA was extracted with TRIzol-up (EZBioscience, China) and 2 µg of RNA were used for reverse transcription with the 4× Reverse Transcription Master Mix (EZBioscience, China). The 2× SYBR Green qPCR Master Mix (EZBioscience, China) was used to perform qRT-PCR. Subsequently, RT-PCR analysis was carried out using the ABI 7500 Fast Real-time PCR system (Applied Biosystems, Switzerland). According to the manufacturer’s instructions, the cycling steps were as follows: reverse transcription (15 min, 42 °C), activation of the Hot-Star DNA Polymerase (5 min, 95 °C), and amplification for 40 cycles (10 s at 95 °C, 40 s at 60 °C). The mRNA level was calculated by 2–ΔΔCt with GADPH as an internal control. Primer sequences are shown in Table 1.

Full table
Western blot analysis
Total protein was extracted from cells with 2% SDS containing a protease inhibitor cocktail (Roche Diagnostics, Roche Applied Science, Meylan, France) on ice for 30 min. Lysates were boiled for 15 min before centrifugation at 14,000 g for 30 min, and the supernatant was collected for protein concentration quantification using a BCA kit (Thermo Scientific, San Jose, CA, USA). A total of 30 mg of proteins were separated using 10% SDS-PAGE gels by electrophoresis and transferred to PVDF membranes. After blocking with 5% skim milk, the membranes were incubated with a primary antibody at room temperature for 3 h and washed with TBST three times. The membranes were then incubated with HRP-conjugated polyclonal goat antibodies (secondary antibodies) at room temperature for 1 h and washed with TBST three times again. Finally, the protein bands were visualized with the TannonTM High-sig ECL Western Blotting Substrate (Tannon, Shanghai, China). GAPDH (Cell Signaling Technology, Danvers, MA, USA) was used as an internal control. The p-gp antibodies (Cell Signaling Technology, Danvers, MA, USA) were diluted 1:1,000 in this study.
Statistical analyses
Results are represented as mean ± standard deviation (SD). Significant differences were analyzed using Student’s t-test and SPSS (version 16.0; SPSS Inc., Chicago, IL, USA) software, where P<0.05 was considered statistically significant. Results were visualized using GraphPad Prism (version 7).
Results
Cisplatin sensitivity and ABCB1 expression in A2780 cells and three cisplatin-resistant variants
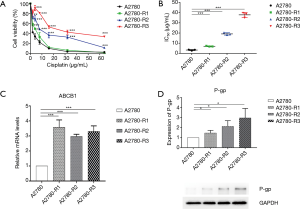
Three cisplatin-resistant variants of the A2780 cells were established during continuous exposure to cisplatin. Fold resistance of three cisplatin-resistant variants was calculated when treated with cisplatin for the 20th, 30th, and 40th cycles, which showed significant differences compared to their parental A2780 cells. The three cisplatin-resistant variants were named A2780-R1, A2780-R2, and A2780-R3, respectively and stored at –80 °C together with A2780 cells. Then, cells were thawed for cytotoxicity analysis and fold resistance calculation was performed. The CCK8 assay was used to evaluate drug resistance of the three cisplatin-resistant variants (Figure 2A). IC50 of the parental A2780 cells was obtained at a concentration of 3.32±0.51 µg/mL, while IC50 for three cisplatin-resistant variants was established at a concentration of 7.00±0.36, 19.15±1.17, and 37.83±2.47 µg/mL, respectively (Figure 2B). Meanwhile, ABCB1 expression in A2780 cells and three cisplatin resistance variants was determined via RT-PCR and Western blotting. The results are shown in Figure 2C,D. Increased expression of ABCB1 indicated the emergence of a resistant phenotype in three cisplatin-resistant variants.
Morphology and growth rate comparison among A2780 cells and three cisplatin resistance variants
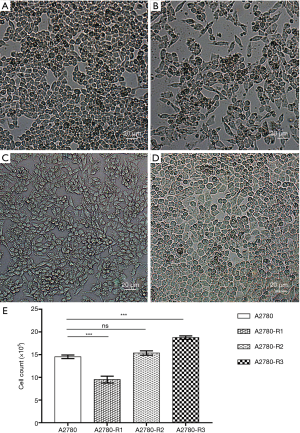
The morphology and growth rates of A2780 cells and three cisplatin-resistant variants were compared by taking photos and counting cells after plating into six-well plates at the same cell density for 48 h (Figure 3A,B,C,D). Compared to parental A2780 cells, A2780-R1 cells appeared flattened and grew slower, A2780-R3 cells were round and grew faster, and A2780-R2 cells had a similar morphology and growth rate (Figure 3E). These data describe the dynamic changes in A2780 cell morphology and growth rates in the process of cisplatin resistance development.
MS analysis of N-glycan profiles in A2780 cells and three cisplatin-resistant variants
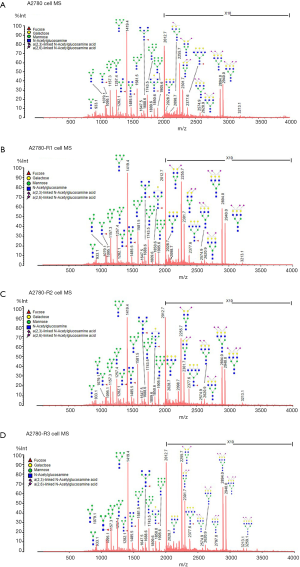
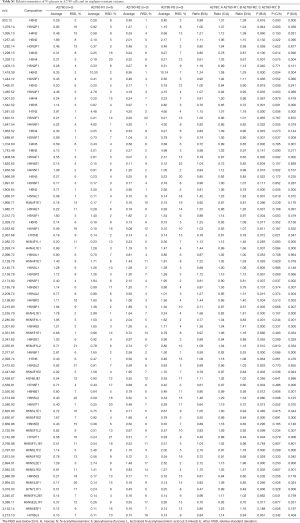
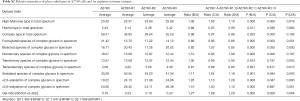
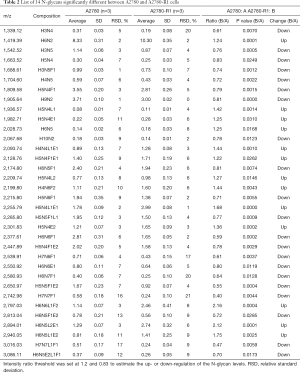
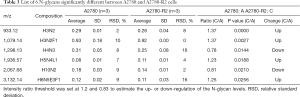
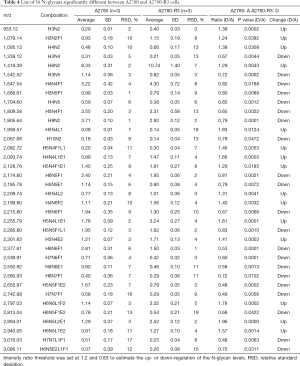
To discover the N-glycan alterations among A2780 cells and three cisplatin-resistant variants, a high-sensitivity method was used for the MALDI MS profiling of N-glycans (18). A total of 77 N-glycans (peaks) were identified and quantified in all samples (Figure 4), including high mannose, hybrid, complex, fucose, bisecting, sialic acid (α2,3- and α2,6-linkages), diantennary, tri-antennary, tetra-antennary, and galactose glycoforms. Relative N-glycan intensities in A2780 cells and three cisplatin-resistant variants are shown in Tables S1,S2,S3. The up- or down-regulation of the N-glycan levels in three cisplatin-resistant variants was estimated and compared to A2780 cells (Tables 2,3,4).
Full table
Full table

Full table
Full table
Full table
Full table
The α2,3-linked sialic acid was significantly increased in all three cisplatin-resistant variants compared to the parental A2780 cells (Figure 5A). Similar increases were also observed in N-glycan gal-ratio (Figure 5B). N-glycan gal-ratio was measured from the relative intensities of agalactosylated (signal at m/z 1,485), monogalactosylated (signal at m/z 1,647), and digalactosylated (signal at m/z 1,809) N-glycans according to the formula G0/(G1+G2×2), which was derived from the IgG gal-ratio reported in our previous work (22). Furthermore, high mannose type glycan was slightly increased in three cisplatin-resistant variants (Figure 5C). The fucosylated glycans were significantly lower in three cisplatin-resistant variants than in parental A2780 cells (Figure 5D). In addition, glyco-subclasses of the α2,6-linked sialic acid, total sialic acid, hybrid, complex, bisecting GlcNAc, diantennary, tri-antennary, and tetra-antennary were analyzed. Results did not show significant differences in all three cisplatin-resistant variants compared to the A2780 cells (Figure 5E,F,G,H,I,J,K,L). Therefore, the results indicated that the levels of α2,3-linked sialic acid, N-glycans gal-ratio, high mannose type glycans, and fucosylated glycans are related to the sensitivity of cisplatin in ovarian cancer A2780 cells. Those glyco-subclasses can be used as biomarkers to monitor the development of platinum resistance.
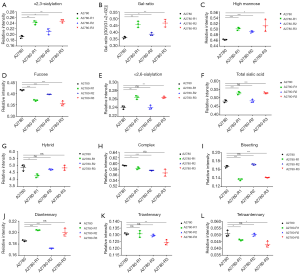
Interestingly, ten glycans displayed significant differences in all three cisplatin-resistant variants compared to A2780 cells, reflecting the dynamic changes in glycans related to cisplatin resistance from A2780 cells to A2780-R3 cells. It was demonstrated that signal at m/z 1,936.6 with α2,3-linked sialic acid was significantly higher in cisplatin-resistant variants as compared to A2780 cells (Figure 6A). Two high-mannose N-glycans (signals at m/z 1,419.4 and 1,581.5) increased and two others (signals at m/z 1,905.6 and 2,067.7) decreased in cisplatin-resistant variants (Figure 6B,C,D,E). However, three fucosylated N-glycans (signals at m/z 1647.5, 1,809.6, and 2,174.8) and two glycans (signals at m/z 1,542.5 and 1,688.6) with bisecting GlcNAc were significantly decreased in cisplatin-resistant variants (Figure 6F,G,H,I,J). The dynamic changes of those glycans can be used to monitor the development of platinum resistance in ovarian cancer A2780 cells.
Variation of glycogene expression in cisplatin-resistant variants of A2780 cells
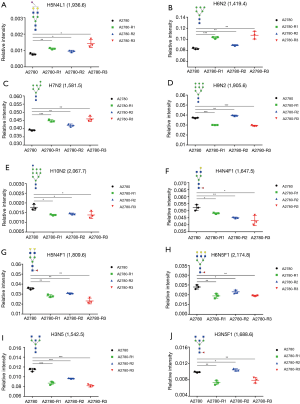
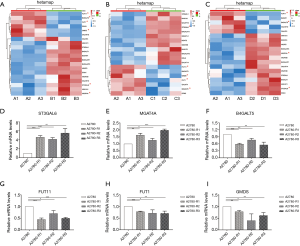
Transcriptome sequencing and analysis were conducted to identify the differences in glycogene expression among A2780 cells and its three cisplatin-resistant variants. Genes involved in N-glycan synthesis that were significantly changed in cisplatin-resistant variants were selected and represented in a heat map (Figure 7A,B,C). In general, transcriptome sequencing results were consistent with the N-glycan alteration analysis. The gene for ST3 beta-galactoside alpha-2,3-sialyltransferase 6 (ST3GAL6), which catalyzes the synthesis of the α2,3-linked sialic acid, has a higher expression in three cisplatin-resistant variants. However, FUT11 and FUT1 genes, which encode alpha-(1,3)-fucosyltransferase and galactoside 2-alpha-L-fucosyltransferase 1, respectively, were down-regulated in three cisplatin-resistant variants. Furthermore, GDP-mannose-4,6-dehydratase (GMDS) gene, an important player in the fucose biosynthesis pathway, was also decreased in three cisplatin-resistant variants. In addition, expression of the B4GALT5 gene was decreased in accordance with an increased N-glycan gal-ratio in three cisplatin-resistant variants. Although tri-antennary and tetra-antennary glycans were not significantly elevated in three cisplatin-resistant variants, the MGAT4A gene encoding alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A was slightly increased in all of them. These results were validated via RT-PCR (Figure 7D,E,F,G,H,I). The higher gene expression of MAN1A1 and MAN1C1, encoding class I mammalian Golgi 1,2-mannosidase, which catalyzes the hydrolysis of three terminal mannose residues from peptide-bound Man [9]-GlcNAc [2] oligosaccharides, was identified in three cisplatin-resistant variants via transcriptome sequencing. However, this result was not in accordance with the RT-PCR analysis as no significant differences in expression were observed between A2780 cells and each cisplatin-resistant variant (data not shown).
Discussion
Ovarian cancer is the second most lethal disease in gynecological malignancies, mainly due to relapse and drug resistance. Most ovarian cancer patients initially respond to platinum compounds and develop platinum resistance following continuous chemotherapy (23). Ovarian cancer patient prognosis after acquiring platinum resistance is extremely poor (10,24). Therefore, it is crucial to conduct studies on the dynamic process during continuous cycles of chemotherapy, which may not only help to develop predictive biomarkers for monitoring platinum resistance, but also provide more information about the mechanisms of platinum resistance and improve patient prognosis. Glycosylation plays an important role in many physiological and pathological processes as well as mechanisms underlying platinum resistance (9,25). There are several glycomic analyses utilized in drug-resistant ovarian cancer. However, none of them concentrate on specific N-glycan changes associated with platinum resistance during continuous cycles of chemotherapy. Furthermore, few glycomic analyses associated with platinum resistance have been performed in cancer cells lines. Cancer cells lines are more readily available and are stable, making them suitable for high-throughput screening. Therefore, this is the first study to identify platinum resistance related to N-glycan changes in ovarian cancer cells during continuous exposure to cisplatin.
The present study mimics the development of platinum resistance in ovarian cancer by continuously exposing A2780 cells to cisplatin. The three cisplatin-resistant variants of the A2780 cells were established after 20, 30, and 40 continuous exposures to cisplatin, making them relatively stable cisplatin-resistant cells. These cells showed significant differences in improved fold resistance compared to the A2780 cells. More cisplatin-resistant variants could not be selected because of death following cisplatin exposure. Then, the higher expression of ABCB1 in cisplatin-resistant variants was identified by RT-PCR and Western-blotting, which indicated the emergence of a resistant phenotype. High-sensitivity MS analysis combined with ethyl esterification derivatization were used to determine N-glycan alterations, including neutral N-glycans and ethyl esterified sialic acid among the four groups, as well as to discriminate between α2,3- and α2,6-linkage N-acetylneuraminic acid. Changes in N-glycan modifications have been mainly attributed to alterations in the corresponding glycosyltransferases (26). Thus, glycogene expression was analyzed using transcriptome sequencing and further validated via RT-PCR. These results were generally consistent with the MS analysis.
This study demonstrated that expression of α2,3-linked sia structures and N-glycan gal-ratio increased, while expression of fucosylation structures decreased in three cisplatin-resistant variants compared to A2780 cells. Many studies have shown that abnormal sialylation is involved in drug resistance. For example, serum levels of α2,3-sialylated glycans were elevated in drug-resistant oral cancer patients compared to drug-sensitive patients (27). Sialylation was probably involved in the development of multidrug resistance in acute myeloid leukemia cells via ST3GAL5 or ST8SIA4 genes (14). Moreover, it has been reported that the ST6GAL1 gene has a significant impact on multidrug resistance by mediating the PI3K/Akt signaling activity and expression of P-gp and MRP1 (28). ST6GAL1 sialyltransferase confers cisplatin resistance by reducing the activation of caspase-3 in ovarian cancer cells (29). To the best of our knowledge, this study demonstrated for the first time that both the expression of ST3GAL6 and α2,3-sialylated glycans were increased in three cisplatin-resistant variants of ovarian cancer cells, suggesting that α2,3-sialylation might be involved in cisplatin resistance via ST3GAL6.
The IgG gal-ratio was used to evaluate the degree of IgG galactosylation and was found to be significantly higher in 12 types of cancer compared to healthy controls, showing great potential for cancer diagnosis as a pan-cancer biomarker (22). Surprisingly, N-glycan gal-ratio in the current study was increased in three cisplatin-resistant variants of A2780 cells, which might be related to lower expression of B4GALT5 gene and be a potential biomarker for platinum resistance.
The N-glycomic analyses in this study have shown that high-mannose structures slightly increased and fucosylation structures significantly decreased in three cisplatin-resistant variants of A2780 cells. RT-PCR results also showed that the FUT11 and FUT1 genes, which catalyze the transfer of the fucose residue to oligosaccharide acceptor in α1, 3/4-, and α1, 2-linkages, were down-regulated in three cisplatin-resistant variants. The lower level of fucosylation structures was further supported by decreased expression of the GMDS gene involved in GDP-L-fucose synthesis in cisplatin-resistant variants (30). In line with these results, Zhao et al. (25) reported that higher expression of high-mannose structures and lower expression of Fuc α1-2(gal β1-4) GlcNAc were identified in the cisplatin-resistant cell line A2780-cp using a lectin blot.
In addition, Alley et al. (16) performed the N-linked glycomic analysis of ovarian cancer serum samples and identified that the level of tri- and tetra-antennary structures with varying degrees of sialylation and fucosylation increased accompanied by decreased levels of bisecting oligosaccharides in late-stage recurrent ovarian cancer patients. However, our MS data suggested that the level of tri- and tetra-antennary were not significantly increased in three cisplatin-resistant variants, although higher gene expression of MGAT4A was identified in three cisplatin-resistant variants compared to parental A2780 cells.
Strikingly, compared to A2780 cells, ten N-glycans exhibited significant differences in all three cisplatin-resistant variants, reflecting their dynamic changes from A2780 cells to A2780-R3 cells. These glycans may play a crucial role in cisplatin resistance and can be used as potential biomarkers to monitor the development of cisplatin resistance in ovarian cancer A2780 cells. For example, the higher level of signal at m/z 1936.6 with α2,3-linked sialic acid and lower level of signal at m/z 1,809.6 with fucosylated glycan might be useful in a clinical setting to monitor the development of platinum resistance.
To the best of our knowledge, this is the first study illustrating N-glycan alterations related to cisplatin resistance in ovarian cancer cells during continuous exposure to cisplatin in vitro. These results have an important clinical significance. On one hand, altered N-glycans may serve as biomarkers for monitoring platinum resistance in ovarian cancer patients during continuous cycles of chemotherapy, deserving further study and validation in patient-derived tumor xenografts or clinical samples (31,32). They can be used to monitor patient response to cisplatin treatment during chemotherapy and guide individualized treatment of ovarian cancer patients in a clinical setting. On the other hand, these altered N-glycans might be involved in cisplatin resistance mechanisms. Further study is needed to elucidate this mechanism and provide a better understanding of platinum resistance.
In conclusion, α2,3-linked sialic structures, found to be increased in cisplatin-resistant variants compared to A2780 cells, might serve as biomarkers to monitor the development of platinum resistance and to guide individualized treatment of ovarian cancer patients.
Acknowledgments
Funding: This work was supported by National Key R&D Program of China (2016YFC1303100) and Natural Science Foundation of China (31770858, 31630088).
Footnote
Conflict of Interests: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethics approval was not required as our experiments only use cell lines.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J Clin 2019;69:280-304. [PubMed]
- Patch AM, Christie EL, Etemadmoghadam D, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015;521:489-94. [Crossref] [PubMed]
- Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284-96. [Crossref] [PubMed]
- Gonzalez Bosquet J, Newtson AM, Chung RK, et al. Prediction of chemo-response in serous ovarian cancer. Mol Cancer 2016;15:66. [Crossref] [PubMed]
- Magalhães A, Duarte HO, Reis CA. Aberrant glycosylation in cancer: a novel molecular mechanism controlling metastasis. Cancer Cell 2017;31:733-5. [Crossref] [PubMed]
- Agrawal P, Fontanals-Cirera B, Sokolova E, et al. A systems biology approach identifies FUT8 as a driver of melanoma metastasis. Cancer Cell 2017;31:804-19.e7. [Crossref] [PubMed]
- Kiermaier E, Moussion C, Veldkamp CT, et al. Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science 2016;351:186-90. [Crossref] [PubMed]
- Reticker-Flynn NE, Bhatia SN. Aberrant glycosylation promotes lung cancer metastasis through adhesion to galectins in the metastatic niche. Cancer Discov 2015;5:168-81. [Crossref] [PubMed]
- Ferreira JA, Peixoto A, Neves M, et al. Mechanisms of cisplatin resistance and targeting of cancer stem cells: Adding glycosylation to the equation. Drug Resist Updat 2016;24:34-54. [Crossref] [PubMed]
- Alkema NG, Wisman GB, van der Zee AG, et al. Studying platinum sensitivity and resistance in high-grade serous ovarian cancer: different models for different questions. Drug Resist Updat 2016;24:55-69. [Crossref] [PubMed]
- Pinho SS, Seruca R, Gartner F, et al. Modulation of E-cadherin function and dysfunction by N-glycosylation. Cell Mol Life Sci 2011;68:1011-20. [Crossref] [PubMed]
- Wojtowicz K, Januchowski R, Nowicki M, et al. Inhibition of protein glycosylation reverses the MDR phenotype of cancer cell lines. Biomed Pharmacother 2015;74:49-56. [Crossref] [PubMed]
- Beretta GL, Benedetti V, Cossa G, et al. Increased levels and defective glycosylation of MRPs in ovarian carcinoma cells resistant to oxaliplatin. Biochem Pharmacol 2010;79:1108-17. [Crossref] [PubMed]
- Ma H, Zhou H, Song X, et al. Modification of sialylation is associated with multidrug resistance in human acute myeloid leukemia. Oncogene 2015;34:726-40. [Crossref] [PubMed]
- Cheng L, Luo S, Jin C, et al. FUT family mediates the multidrug resistance of human hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell Death Dis 2013;4:e923. [Crossref] [PubMed]
- Alley WR, Vasseur JA, Goetz JA, et al. N-linked glycan structures and their expressions change in the blood sera of ovarian cancer patients. J Proteome Res 2012;11:2282-300. [Crossref] [PubMed]
- Holst S, Heijs B, de Haan N, et al. Linkage-specific in situ sialic acid derivatization for N-glycan mass spectrometry imaging of formalin-fixed paraffin-embedded tissues. Anal Chem 2016;88:5904-13. [Crossref] [PubMed]
- Holst S, Wilding JL, Koprowska K, et al. N-glycomic and transcriptomic changes associated with CDX1 mRNA expression in colorectal cancer cell lines. Cells 2019. [Crossref] [PubMed]
- McDermott M, Eustace AJ, Busschots S, et al. In vitro development of chemotherapy and targeted therapy drug-resistant cancer cell lines: a practical guide with case studies. Front Oncol 2014;4:40. [Crossref] [PubMed]
- Yan XD, Pan LY, Yuan Y, et al. Identification of platinum-resistance associated proteins through proteomic analysis of human ovarian cancer cells and their platinum-resistant sublines. J Proteome Res 2007;6:772-80. [Crossref] [PubMed]
- Vreeker GCM, Nicolardi S, Bladergroen MR, et al. Automated plasma glycomics with linkage-specific sialic acid esterification and ultrahigh resolution MS. Anal Chem 2018;90:11955-61. [Crossref] [PubMed]
- Ren S, Zhang Z, Xu C, et al. Distribution of IgG galactosylation as a promising biomarker for cancer screening in multiple cancer types. Cell Res 2016;26:963-6. [Crossref] [PubMed]
- Shin DH, Kwon GS. Pre-clinical evaluation of a themosensitive gel containing epothilone B and mTOR/Hsp90 targeted agents in an ovarian tumor model. Journal of Controlled Release 2017;268:176-83. [Crossref] [PubMed]
- El Bairi K, Amrani M, Kandhro AH, et al. Prediction of therapy response in ovarian cancer: where are we now? Crit Rev Clin Lab Sci 2017;54:233-66. [Crossref] [PubMed]
- Zhao R, Qin W, Qin R, et al. Lectin array and glycogene expression analyses of ovarian cancer cell line A2780 and its cisplatin-resistant derivate cell line A2780-cp. Clin Proteomics 2017;14:20. [Crossref] [PubMed]
- Kizuka Y, Taniguchi N. Enzymes for N-glycan branching and their genetic and nongenetic regulation in cancer. Biomolecules 2016. [Crossref] [PubMed]
- Shah MH, Telang SD, Shah PM, et al. Tissue and serum alpha 2-3- and alpha 2-6-linkage specific sialylation changes in oral carcinogenesis. Glycoconj J 2008;25:279-90. [Crossref] [PubMed]
- Ma H, Cheng L, Hao K, et al. Reversal effect of ST6GAL 1 on multidrug resistance in human leukemia by regulating the PI3K/Akt pathway and the expression of P-gp and MRP1. PLoS One 2014;9:e85113. [Crossref] [PubMed]
- Schultz MJ, Swindall AF, Wright JW, et al. ST6Gal-I sialyltransferase confers cisplatin resistance in ovarian tumor cells. J Ovarian Res 2013;6:25. [Crossref] [PubMed]
- Holst S, Deuss AJ, van Pelt GW, et al. N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (CDX1)/villin mRNA expression. Mol Cell Proteomics 2016;15:124-40. [Crossref] [PubMed]
- Cornelison R, Dobbin ZC, Katre AA, et al. Targeting RNA-polymerase I in both chemosensitive and chemoresistant populations in epithelial ovarian cancer. Clin Cancer Res 2017;23:6529-40. [Crossref] [PubMed]
- Boone JD, Dobbin ZC, Straughn JJ, et al. Ovarian and cervical cancer patient derived xenografts: The past, present, and future. Gynecol Oncol 2015;138:486-91. [Crossref] [PubMed]


