
Cell adhesion molecule 2 (CADM2) promotes brain metastasis by inducing epithelial-mesenchymal transition (EMT) in human non-small cell lung cancer
Introduction
Brain metastasis is the most feared complication of cancer and the most common intracranial tumor in adults (1). Lung cancer is one of the most common tumors among men and women worldwide, and 85% of them are non-small cell lung cancer (NSCLC) (2,3). Up to 50% of NSCLC patients are expected to experience brain metastasis at some point (4). The mechanism of tumor brain metastasis is still unclear, and due to the migration and invasiveness of NSCLC, the existing treatment for NSCLC in patients with brain metastasis is generally ineffective, the prognosis is reduced, and the median survival time is only a few months (5). Finding molecular targets, understanding the mechanisms of brain metastasis, and discovering new diagnostic markers or therapeutic targets are particularly urgent.
In advanced NSCLC, localized large mediastinal lymph nodes (>2 cm) (6), non-squamous cell carcinoma (7), and age ≤60 years were found to be associated with high brain metastasis rates. However, we currently know little about the genetic and genomic changes that occur in NSCLC brain metastasis. Through tumor genomics and proteomics methods, we can now study the above changes as a whole to comprehensively understand the changes leading to brain metastasis in transcription levels, exploring the molecular mechanism of NSCLC with brain metastasis, and possibly identifying new therapeutic targets.
In this study, we found that cell adhesion molecule 2 (CADM2) is up-regulated in NSCLC patients with brain metastasis compared to NSCLC patients without brain metastasis. CADM2 is a member of the synaptic cell adhesion molecule one family that is involved in homologous and heterophilic interactions with other members of the Nectin-like family, leading to cell aggregation (8,9). In renal cell carcinoma (10) and prostate cancer (11), CADM2 acts as a tumor suppressor. Low CADM2 expression in patients with hepatocellular carcinoma predicts a higher risk of recurrence after hepatectomy (12). However, we found that silencing of CADM2 inhibited cell migration, invasion, and epithelial-mesenchymal transition (EMT) in NSCLC cell lines A549 and H322, indicating that CADM2 may be a poor prognostic factor for NSCLC patients with brain metastasis.
Methods
Subjects
Tissues used in this study were obtained from the Affiliated Cancer Hospital & Institute of Guangzhou Medical University, including samples of NSCLC with [n=26, brain metastasis (BM) group] or without (n=36, nonBM group) brain metastasis. We received ethical approval from the local research ethics committee, and all subjects received written informed consent for sample collection and research use. All diagnoses of NSCLC were confirmed by radiographic imaging and other methods, of those cases that brain metastases were observed.
Microarray and data analysis
Trizol/Chloroform was used to extract total RNA from samples, and Affymetrix Human Transcriptome Array 2.0 analysis (Affymetrix, Santa Clara, CA, USA) was performed by Gene Tech Company Limited (Shanghai, China). Briefly, after RNA measurement using the Nanodrop ND-1000 and denaturing gel electrophoresis, the samples were amplified and labeled with biotin by terminal deoxynucleotidyl transferase (TdT). Labeled DNA was then hybridized with the Affymetrix GeneChip™ Human Transcriptome Assay 2.0 (Cat No. 902309). Slides were carefully washed and scanned with Affymetrix® GeneChip Command Console (AGCC) which installed in GeneChip® Scanner 3000 7G. The data were analyzed with Robust Multichip Analysis (RMA) algorithm. Normalized data were further analyzed using moderated t-test. Differentially expressed genes were named through fold-change (fold change >2, P<0.05) screening.
Quantitative real-time PCR (qRT-PCR)
Total RNA from lung tissue was extracted using E.Z.N.A.TM Fast Protocol for FFPE Tissue (OMEGA, Guangzhou, China), and total RNA from cell samples was extracted using TRIzol Reagent (Invitrogen) following the manufacturer’s protocols. Then the extracted RNA was reverse-transcribed into cDNA, using the Prime ScriptTMRT reagent Kit (TaKaRa, Shiga, Japan). qRT-PCR was performed using Premix EX TaqTM (Probe qPCR) (TaKaRa). The primers used are as follows:
Forward primer: 5'- GCTCTGGGCCTCATGGTTT-3'
Reverse primer: 5'-CAGCTGAGCAGAGGCAACTTT-3'
Probe primer: 5'- FAM-TTCTCTTGGAGCCTTTCATTACAGA-BHQ1-3'.
Amplification was performed using the following PCR profile: preheating at 94 °C for 3 min, followed by 40 cycles of 94 °C for 3 min, 94 °C for 5 s, and 60 °C for 30 s. PCR reactions were performed on an ABI PRISM® 7300 Sequence Detection System (Foster City, CA, USA). Gene expression was measured in triplicate, quantified by the 2−ΔΔCt method (13), and normalized using GAPDH as the internal control.
Cell culture and siRNA transfection
NSCLC cell lines A549 and H322 were bought from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone). The negative control siRNA (si-NC) and siRNAs targeting CADM2 (si-CADM2) were bought from GenePharma (Suzhou, China). Cells were plated to 50% confluency and transfected with 10, 50, and 100 nM si-NC or si-CADM2, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The sequences of si-CADM2 are:
5'-UUCAACAACGGUUACAUUCUG-3' (sense)
5'-GAAUGUAACCGUUGUUGAAGG-3' (antisense)
Transwell assays
Transwell assay was carried out to detect the migration and invasion abilities of A549 and H322 after transfection with si-NC or si-CADM2. For migration, transfected A549 and H322 cells were harvested, and 1×105 cells in 100 µL of 0.1% serum medium inserted in the upper chamber (pore size, 8 µm) (Becton Dickinson Labware). The lower chamber was filled with 600 µL DMEM (Hyclone) supplemented with 10% FBS (Hyclone). For invasion, the same density of cells was placed into the upper chambers, which were pre-coated with Matrigel (BD Biosciences). After incubation for 24 hours, the cells in the upper chamber of the filter are removed with a cotton swab, the cells on the underside were fixed with 4% paraformaldehyde for 15 min, stained with 0.1% crystal blue for 10 min. The migrated or invasive cells were counted by photographing at 200× magnification with LEICA Microscope (Tokyo, Japan) in five independent fields for each well. The assays were performed in triplicate.
Western blot analysis
Total protein from cell samples was extracted using RIPA Lysis Buffer (Beyotime Biotechnology, Shanghai, China). Thirty micrograms of protein were separated by 10% SDS polyacrylamide gel electrophoresis and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). Membranes were then blocked with 5% non-fat milk for 1 h at 37 °C. Subsequently, membranes were incubated in 5% non-fat milk for one h at 37 °C with anti-CADM2, anti-Vimentin, anti-E-cadherin, and anti-GAPDH primary antibodies. After washing with TBST (TBS holding 0.5% Tween 20), membranes were incubated at 37 °C for 40 min with an HRP-conjugated secondary antibody. Membranes were then rewashed with TBST and assayed via enhanced chemiluminescence (ECL) and recorded on X-ray films.
Statistical analysis
Statistical analysis was conducted using SPSS 19.0 software package (SPSS Inc., Chicago, IL, USA). Student’s t-test was used for comparisons between two groups, and one-way ANOVA was used for multiple comparisons. A value of P<0.05 was considered statistically significant.
Results
Expression of CADM2 in NSCLC with or without brain metastasis patients
Human transcriptome array analysis and qRT-PCR were performed to determine the expression of CADM2 in NSCLC with or without brain metastasis patients. Compared with NSCLC patients without brain metastasis (nonBM), 513 up-regulated, and 438 down-regulated genes were found in NSCLC plus BM patients, data were shown in http://fp.amegroups.cn/cms/cd0b98a53c8f6fa0f12bc5c789fd6322/atm.2020.03.85-1.pdf. Figure 1A was the heatmap of the up-regulated and down-regulated genes. From this, we found that CADM2 was up-regulated in BM patients compared to nonBM patients. The gene expression level of CADM2 was verified in 26 BM samples and 36 nonBM samples using qRT-PCR assays. Table 1 shows the baseline characteristics of the test subjects. As a result, shown, CADM2 was up-regulated 2.05-fold in BM patients compared to nonBM patients (Figure 1B). And there was a good match between the result of microarray and qRT-PCR (Figure 1C).
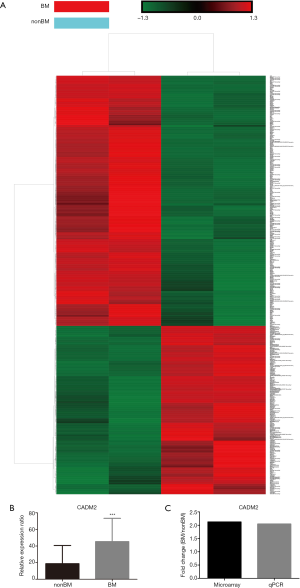
Full table
Silencing of CADM2 by siRNAs
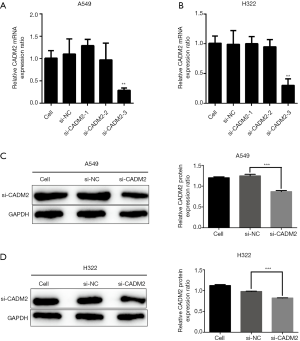
To reduce the expression of CADM2, we synthesized NC and three active siRNAs targeting different CADM2 sequences and then transfected A549 and H322 cells. qRT-PCR and western blot assays were used to determine the efficiency of the silencing. In the siRNAs evaluated, the lowest expression of CADM2 mRNA in A549 and H322 cells was observed by the transfection of si-CADM2-3 (Figure 2A,B) and it also proved to be able to successfully knockdown CADM2 protein (Figure 2C,D).
Silencing of CADM2 inhibited cell migration and invasion in NSCLC cells
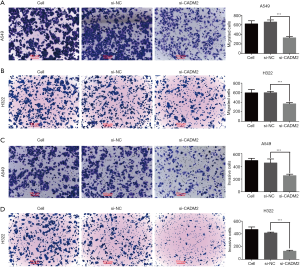
Transwell assay was performed following the transfection of si-CADM2 or si-NC to prove the role of CADM2 in regulating cell migration and invasion in NSCLC cells. The results showed that the number of cells entering the lower chamber through the membrane of si-CADM2-transfected A549 and H322 cells was significantly lower than that of si-NC transfected cells (Figure 3A,B), indicating that silencing of CADM2 inhibited cell migration in NSCLC cells. Besides, the number of cells that passed through the matrigel-coated membrane into the lower chamber was significantly lower in the si-CADM2 transfected A549 and H322 cells than in the si-NC transfected A549 and H322 cells (Figure 3C,D), indicating that silencing of CADM2 inhibited cell invasion in NSCLC cells.
Silencing of CADM2 inhibited EMT in NSCLC cells
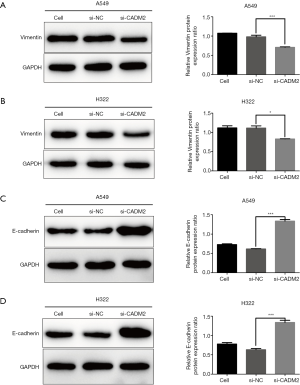
The protein expression levels of Vimentin and E-cadherin were detected after transfection with si-CADM2 or si-NC to prove the role of CADM2 in regulating NSCLC cell EMT. The results showed that CADM2 knockdown significantly decreased the level of Vimentin (mesenchymal marker) (Figure 4A,B), and correspondingly increased the protein level of the E-cadherin (epithelial marker) in both A549 and H322 cells (Figure 4C,D).
Discussion
Brain metastasis is a common complication of NSCLC, which is associated with severe morbidity and mortality. Understanding the cellular and molecular mechanisms of lung cancer-related brain metastasis will supply hope for effective treatment and improve the prognosis of the disease. Preventive treatment may prevent or delay brain metastasis; showing high-risk patients for brain metastasis may be a better option for those who are at high risk. It is reported that some proteins or signaling pathways take part in brain metastasis of lung cancer. The expressions of CDH2 (N-cadherin), KIFC1, and FALZ, are highly predictive for brain metastasis in early and advanced lung cancer and can be used to identify brain metastasis high-risk patients (14). Also, the ability of lung adenocarcinoma cells to metastasize to the brain was enhanced by Wnt/Tcf signal program of LEF1 and HOXB9 (15), and recently, Claudin-5 (16), ACTN4 (17) and ADAM9 (18) have been proved to be associated with lung cancer metastasis. However, the mechanism of brain metastasis of NSCLC is still unclear, so more research is needed. In the current study, we use human genome array analysis to study the gene profiles of NSCLC patients with or without brain metastasis, to identify genes that contribute to brain metastasis of NSCLC patients. Our research supplies a comprehensive understanding of the changes in transcriptional levels of brain metastasis in NSCLC, and we also supply information on the potential cellular and molecular mechanisms.
To our knowledge, this is the first study investigating CADM2 in NSCLC with brain metastasis. Based on microarray analysis and qRT-PCR, it was showed that CADM2 was up-regulated more than two folds in NSCLC patients with brain metastasis compared to those without brain metastasis. Further study showed that the silence of CADM2 inhibited the migration and invasion of NSCLC cell lines A549 and H322. These results showed that CADM2 might play a key role in preventing brain metastasis in NSCLC. However, it has been reported that overexpression of CADM2 may inhibit cell proliferation in the prostate, renal, and endometrial cancers (7,10,11). Also, the overexpression of CADM2 could promote apoptosis and inhibit cell invasion in renal and endometrial cancer cells (10). We must do more research to clarify this opposite effect of CADM2 in different cancers.
Furthermore, we found that CADM2 knockdown markedly reduced the level of Vimentin, a mesenchymal marker, and increased the level of E-cadherin, an epithelial marker, suggesting that CADM2 could promote EMT in NSCLC. EMT is a conservative evolutionary process and is considered necessary for normal embryonic development (6). EMT is an essential biological process in the development of malignant tumors, such as enhancing the invasion and metastasis of cancer cells (19-21). Also, increased studies have shown that EMT (22-24) can regulate the ability of cancer cells to migrate and invade. Based on these reports, we speculate that CADM2 promotes brain metastasis by inducing EMT in human NSCLC.
Our study focused on the mechanism of CADM2 in NSCLC with brain metastasis patients and showed that CADM2 might promote brain metastasis in NSCLC patients by inducing EMT. In conclusion, CADM2 may be a new target for the prevention and treatment of brain metastasis in NSCLC soon.
Acknowledgments
Funding: The project was supported by the National Natural Science Foundation of China (No. 81572258).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.85). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. We received ethical approval from the local research ethics committee, and all subjects received written informed consent for sample collection and research use.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maher EA, Mietz J, Arteaga CL, et al. Brain metastasis: opportunities in basic and translational research. Cancer Res 2009;69:6015-20. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Rykova EY, Ponomaryova AA, Zaporozhchenko IA, et al. Circulating DNA-based lung cancer diagnostics and follow-up: looking for epigenetic markers. Transl Cancer Res 2018;7:S153-70. [Crossref]
- Nolte SM, Venugopal C, McFarlane N, et al. A cancer stem cell model for studying brain metastases from primary lung cancer. J Natl Cancer Inst 2013;105:551-62. [Crossref] [PubMed]
- Govindan R, Page N, Morgensztern D, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol 2006;24:4539-44. [Crossref] [PubMed]
- Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871-90. [Crossref] [PubMed]
- He Z, Xu H, Meng Y, et al. miR-944 acts as a prognostic marker and promotes the tumor progression in endometrial cancer. Biomed Pharmacother 2017;88:902-10. [Crossref] [PubMed]
- Fogel AI, Akins MR, Krupp AJ, et al. SynCAMs organize synapses through heterophilic adhesion. J Neurosci 2007;27:12516-30. [Crossref] [PubMed]
- Biederer T. Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics 2006;87:139-50. [Crossref] [PubMed]
- He W, Li X, Xu S, et al. Aberrant methylation and loss of CADM2 tumor suppressor expression is associated with human renal cell carcinoma tumor progression. Biochem Biophys Res Commun 2013;435:526-32. [Crossref] [PubMed]
- Chang G, Xu S, Dhir R, et al. Hypoexpression and epigenetic regulation of candidate tumor suppressor gene CADM-2 in human prostate cancer. Clin Cancer Res 2010;16:5390-401. [Crossref] [PubMed]
- Yang S, Yan HL, Tao QF, et al. Low CADM2 expression predicts high recurrence risk of hepatocellular carcinoma patients after hepatectomy. J Cancer Res Clin Oncol 2014;140:109-16. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Grinberg-Rashi H, Ofek E, Perelman M, et al. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res 2009;15:1755-61. [Crossref] [PubMed]
- Nguyen DX, Chiang AC, Zhang XH, et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell 2009;138:51-62. [Crossref] [PubMed]
- Ma SC, Li Q, Peng JY, et al. Claudin-5 regulates blood-brain barrier permeability by modifying brain microvascular endothelial cell proliferation, migration, and adhesion to prevent lung cancer metastasis. CNS Neurosci Ther 2017;23:947-60. [Crossref] [PubMed]
- Gao Y, Li G, Sun L, et al. ACTN4 and the pathways associated with cell motility and adhesion contribute to the process of lung cancer metastasis to the brain. BMC Cancer 2015;15:277. [Crossref] [PubMed]
- Lin C, Chen H, Huang C, et al. ADAM9 promotes lung cancer metastasis to brain by a plasminogen activator-based pathway. Cancer Res 2014;74:5229-43. [Crossref] [PubMed]
- Jiang Z, Sun Y, Wang S, et al. Epithelial-mesenchymal transition: potential regulator of ABC transporters in tumor progression. J Cancer 2017;8:2319-27. [Crossref] [PubMed]
- Fedele M, Cerchia L, Chiappetta G. The Epithelial-to-Mesenchymal Transition in Breast Cancer: Focus on Basal-Like Carcinomas. Cancers (Basel) 2017. [Crossref] [PubMed]
- Zaman A, Bivona TG. Emerging application of genomics-guided therapeutics in personalized lung cancer treatment. Ann Transl Med 2018;6:160. [Crossref] [PubMed]
- Chen D, Zhou H, Liu G, et al. SPOCK1 promotes the invasion and metastasis of gastric cancer through Slug-induced epithelial-mesenchymal transition. J Cell Mol Med 2018;22:797-807. [Crossref] [PubMed]
- Wang W, Zhao Y, Yao S, et al. Nigericin Inhibits Epithelial Ovarian Cancer Metastasis by Suppressing the Cell Cycle and Epithelial-Mesenchymal Transition. Biochemistry Mosc 2017;82:933-41. [Crossref] [PubMed]
- Yang J, Chen Z, Wang X, et al. Inactivation of miR-100 combined with arsenic treatment enhances the malignant transformation of BEAS-2B cells via stimulating epithelial -mesenchymal transition. Cancer Biol Ther 2017;18:965-73. [Crossref] [PubMed]


