
Dexmedetomidine promotes breast cancer cell migration through Rab11-mediated secretion of exosomal TMPRSS2
Introduction
Breast cancer is one of the most common types of cancer affecting women worldwide (1). Surgery is currently the primary approach in breast cancer treatment. However, owing to the high rates of recurrence and metastasis, some patients with breast cancer still have a poor prognosis after treatment (1). Dexmedetomidine (DEX) is a new generation α2-adrenergic receptor agonist that is highly selective and serves as a sedative and an adjuvant to anesthetic strategies in the perioperative context (2). By binding to α2-adrenergic receptors, DEX plays an important role in various biological processes (3). In tumors of a more malignant phenotype characteristic, α2-adrenergic receptor is overexpressed and, in breast cancer, is associated with relapse and poor clinical outcome (4,5). Recent studies showed that DEX increased the proliferation, migration and invasion ability of breast cancer cells in vitro, possibly by regulating the α2-adrenergic receptor/Erk1/2 signaling pathway (6). An in vivo study showed that DEX mainly increased tumor-cell retention and metastasis in mammary adenocarcinoma in rats via α2-adrenergic receptors (7). Interestingly, a prospective randomized clinical study that used breast cancer cell line MCF-7 showed that the postoperative serum of patients who received 2 µg/kg DEX for two hours during surgery but did not receive saline, showed significantly higher proliferation, migration, and invasion compared to serum taken preoperatively (8). This study provides indirect evidence indicative of the possibility of deleterious effects of the perioperative utilization of DEX in the prognosis of breast cancer. Nevertheless, the underlying mechanisms by which DEX promotes breast cancer cell migration remain elusive.
Type II transmembrane serine protease (TTSP) plays a key role in tumor growth, invasion, and metastasis (9). There is a significant association between genetic variants of TTSP genes and the risk and prognosis of breast cancer (10). A member of the TTSP subfamily, transmembrane protease serine 2 (TMPRSS2) is regulated by androgens and is strongly related to prostate cancer (11). In addition, a correlation exists between TMPRSS2 protein levels and the progression of prostate cancer (12). By activating matriptase and disrupting the extracellular matrix (ECM), TMPRSS2 can promote the growth, invasion, and metastasis of prostate cancer cells in vitro (13). TMPRSS2 is expressed as a 70-kDa full-length protein and a 32-kDa cleaved protease domain. The cleaved protease domain of TMPRSS2 is thought to be secreted into cell media after autocleavage (14). Notably, the expression of TMPRSS2 in both its full-length and secreted forms was detected in breast cancer cell line MCF-7 (15). Meanwhile, the influence of TMPRSS2 on the migration of DEX-treated breast cancer cells is still unclear. We hypothesized that DEX may regulate the migration of breast cancer cells through the upregulation of TMPRSS2.
Methods
Antibodies
The following commercial primary antibodies were obtained: rabbit monoclonal anti-TMPRSS2, rabbit anti-heat shock protein 70 (Hsp70), rabbit anti-a disintegrin and metalloproteinase domain-containing protein 10 (ADAM10), rabbit anti-EEA1, rabbit anti-collagen IV, rabbit anti-matrix metallopeptidase 16 (MMP16) and rabbit anti-tenascin C antibodies from Abcam (Cambridge, MA, USA); rabbit anti-α2-adrenergic receptor and mouse anti-Hsp90 antibodies from Sigma-Aldrich (St. Louis, MO, USA); mouse anti-signal transducer and activator of transcription 3 (STAT3), mouse anti-GAPDH, mouse anti-histone H3, rabbit anti-Rab35, rabbit anti-fibronectin, rabbit anti-phospho-STAT3Tyr705, rabbit anti-Rab7, rabbit anti-Rab4 and rabbit anti-Rab11 antibodies from Cell Signaling (Danvers, MA, USA); mouse anti-Rab11 antibody from BD Biosciences (San Jose, CA, USA).
Cell culture and treatment
Human breast adenocarcinoma cell lines MCF-7 and MDA-MB-231 were obtained from ATCC (ATCC, Manassas, VA, USA) and maintained and stored at the College of Life Science of Northeast Agricultural University (Harbin, China). The MCF-7 and MDA-MB-231 cells were cultured in Dulbecco’s Modified Eagle Medium (Gibco/Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (cat. no. 16000–044; Gibco/Thermo Fisher Scientific) and 1% penicillin-streptomycin (cat. no. 15140–122; Gibco/Thermo Fisher Scientific) at 37 °C in a humidified atmosphere of 5% CO2.
Cells were treated with DEX (Sigma-Aldrich) at different concentrations for various periods of time (0, 12, 24 and 48 h). The concentrations of DEX ranged from 0.01 to 1 µM, based on previous studies (6,16-18). In some cases, the cells were pretreated for 1 h with STAT3 inhibitor III (WP1066, cat. no. 573097; EMD Millipore, Kankakee, IL, USA) before DEX was added. They were subsequently incubated for the indicated time periods with or without WP1066.
Knockdown assay
Two validated commercial siRNAs specifically targeted against different coding sites of human TMPRSS2 were used to knockdown TMPRSS2 expression (siTMP-a, cat. no. SASI_Hs01_00072210; siTMP-b, cat. no. SASI_Hs01_00072211; Sigma-Aldrich). Simultaneously, Rab11a and Rab11b isoforms were knocked down using siRNAs targeted against human Rab11a (5'-ugucagacagacgcgaaaa-3') and Rab11b (5'-gcaccugaccuaugagaac-3'), respectively. MISSION® siRNA Universal Negative Control #1 (siCTL, cat. no. SIC-001; Sigma-Aldrich) was used as the control. The siRNAs were introduced at the final concentration of 20 nM with RNAiMAX (cat. no. 13778-075; Invitrogen/Thermo Fisher Scientific). After 48 h, the cells were harvested.
Reverse transcription and real time polymerase chain reaction (PCR)
Total RNA was extracted using Trizol reagent (cat. no. 15596026; Invitrogen) according to the manufacturer’s instructions. SuperScript™ III First-Strand Synthesis Kit (cat. no. 11752-050; Invitrogen) was used to reverse 2 µg of RNA to the complementary DNA (cDNA). The PCR reaction included 1 × SYBR Green PCR Master Mix (cat. no. 1725270; Bio-Rad, Hercules, CA, USA), 2 µL of cDNA, and 0.2 µM of the specific primer pairs. The primers for TMPRSS2 are forward 5'-gtgacagtggagggcctctgg-3' and reverse 5'-cgtcaaggacgaagaccatgt-3', respectively. The primers for GAPDH are forward 5'-tgtgtccgtcgtggatctga-3' and reverse 5'-cctgcttcaccaccttcttga-3', respectively. Real time PCR was performed on a 7500 fast Real-Time PCR system (Applied Biosystems/Thermo Fisher Scientific) with a two-step PCR program: 1 cycle of 10 min at 95 °C; 40 cycles of 15 sec at 95 °C, 45 sec at 56 °C, 30 sec at 72 °C. The relative expression levels of TMPRSS2 were calculated using the 2−ΔΔCt method and normalized to the house-keeping gene GAPDH. The fold change over the controls was presented and compared.
Scratch assay
The migration ability of the MCF-7 and MDA-MAB-231 cells was assessed by performing scratch assay, as described in the literature. Cells were seeded in 6-well plates and cultured when they reached 90% confluence. A scratch or wound was made by scraping a 200 µL sterile plastic pipette tip across the surface of the cell layers. The cells were rinsed with media three times, and then maintained in a fresh serum free medium for 24 or 48 h. The wound was photographed in three randomly selected fields under a phase contrast microscope (Olympus, Tokyo, Japan) at 0 h and at 48 h after the scratch. The experiment was carried out in triplicate, and the migration rate was calculated and compared between different conditions.
ELISA assay
Cell culture media were collected and centrifuged for 10 min at 12,000 rpm at 4 °C. The supernatant was collected and 100 µL was used for measurement of TMPRSS2 with Human TMPRSS2 ELISA Kit (cat. no. GWB-KBBFP3; Genway Biotech Inc., San Diego, CA, USA) according to the manufacturer’s protocol.
Ultracentrifugation: isolation of exosomes
Exosomes were isolated from the cell culture media. Briefly, the media were harvested from treated MCF-7 cells, and then centrifuged in a Beckman Coulter Allegra X-15R centrifuge at 3,000 g at 4 °C for 20 min to remove any detached cells. The supernatant was collected and centrifuged in a Beckman Coulter Optima L-80XP Ultracentrifuge at 10,000 g at 4 °C for 90 min with a Type 50.2 Ti rotor to remove contaminating apoptotic bodies, microvesicles, and cell debris. The clarified media were then centrifuged in a Beckman Coulter Optima L-80XP Ultracentrifuge at 120,000 g at 4 °C for 120 min with a Type 50.2 Ti rotor to pellet exosomes. The supernatant was carefully removed, and the crude exosome-containing pellets were resuspended in ice-cold PBS.
For transmission electron microscopy, the samples were fixed with 4% glutaraldehyde-formalin, and then sent to Northeast Agriculture University Microscopy Research Laboratory Core (Harbin, China) for preparation. The exosomes were examined and imaged with a JEOL 1010 electron microscope (Peabody, MA, USA).
Western blot
Cells were lysed in RIPA buffer [25 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1% NP-40] supplemented with 1× protease and phosphatase inhibitor (cat. no. 78441; Thermo Fisher Scientific). After centrifugation at 4 °C for 20 min, the supernatant was carefully collected and quantitated, before being stored at −80 °C for further analysis. Nuclear protein was isolated using the Nuclear Extraction Kit (cat. no. ab113474; Abcam). Equal amounts of proteins and exosomes were loaded onto 7.5 or 12.5% SDS polyacrylamide gel, and then transferred to a nitrocellulose membrane (cat. no. 88025; Thermo Fisher Scientific). The membrane was blocked for 1 h in 5% nonfat milk or bovine serum albumin, and then incubation with the indicated primary antibodies took place over-night at 4 °C. After incubation with HRP-conjugated goat anti-rabbit or goat anti-mouse IgG (cat. no. G-21234 and G-21040; Invitrogen) for 1 h at room temperature, the signals were detected using ECL Western Blotting Substrate (cat. no. 32109; Pierce/Thermo Fisher Scientific). To remove the primary antibodies for reblotting with the other primary antibody, Stripping Buffer for Western blot (Thermo Fisher Scientific) was applied to some blots. The specific bands were quantitated using Image J (Version 1.51s; National Institute of Health, Rockville, MD, USA).
Indirect immunofluorescence
Cells were cultured on cover slides and treated as described in the literature. The cells were then fixed in 4% paraformaldehyde for 15 min at room temperature, washed 3 times, and then permeabilized with 0.1% Triton X-100 for 10 min. Non-specific staining was blocked with 10% goat serum. The fixed cells were then incubated with rabbit anti-TMPRSS2 antibody (1:100) alone or rabbit anti-TMPRSS2 (1:100) and mouse anti-Rab11 (1:100) antibodies overnight at 4 °C. They were then incubated with the secondary antibodies, Alexa Fluor 488-conjugated goat anti-rabbit IgG (1:2,000; cat. no. A27034, Invitrogen) and Alexa Fluor 594-conjugated goat anti-mouse IgG (1:2,000; cat. no. A11005, Invitrogen), for 1 h at room temperature. The secondary antibodies alone were used as the blank control. The cover slides were mounted with ProLong Gold Antifade Mountant with 4',6-diamidino-2-phenylindole (DAPI; cat. no. P36935, Invitrogen), and images were captured with Zeiss LSM 710 laser scanning confocal system equipped with a 63x oil immersion objective lens (Zeiss, Beijing, China).
Statistical analysis
Data are shown as mean ± SD. Differences between two groups were compared with unpaired t-test and the Mann-Whitney test. One-way ANOVA with the Tukey test and two-way ANOVA with the Sidak correction were used to draw multiple comparisons, respectively. GraphPad Prism 6.0 (La Jolla, CA, USA) was used to perform statistical analysis. Statistical difference was indicated when P<0.05.
Results
TMPRSS2 is upregulated in DEX-treated breast cancer cells
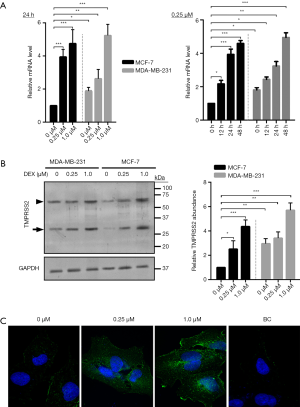
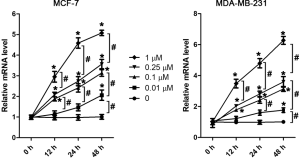
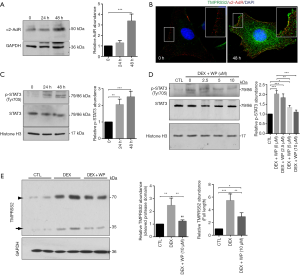
TMPRSS2 is expressed in various human cancer cells, particularly in cells from cancers such as prostate, head, and neck cancer (15). In this study, we hypothesized that if TMPRSS2 is expressed in cultured breast cancer cells in vitro, the regulation of TMPRSS2 by DEX may in turn be demonstrated to regulate the migration and metastasis of breast cancer cells. To test our hypothesis, the expression of TMPRSS2 was firstly assessed in cultured MCF-7 and MDA-MB-231 cells, two of the most commonly used human breast cancer cell lines (19). The results from real-time PCR demonstrated that compared to the non-treated cells, the mRNA levels of TMPRSS2 were significantly upregulated in both a time- and dose-dependent manner in the DEX-treated MCF-7 and MDA-MB-231 cells (Figure 1A, Figure S1). Similar results were found with Western blot assay, which demonstrated a significant increase of TMPRSS2 at the protein levels by DEX (Figure 1B). Image analysis identified two specific bands, with one close to 75 kDa and the other between 37 and 25 kDa (Figure 1B). As previously mentioned, TMPRSS2, a transmembrane serine protease, is expressed in two forms, as a full-length protein and as a cleaved protease domain; which are recognized by 70- and 32-kDa, respectively (14). In our study, an increase in TMPRSS2 in both its full-length and cleaved protease domain forms was detected in DEX-treated MCF-7 and MDA-MB-231 cells (Figure 1B). Notably, we detected higher levels of TMPRSS2 in the non-treated MDA-MB-231 cells than in the non-treated MCF-7 cells (Figure 1A,B). It has been reported that the migration rate of MCF-7 is lower than that of MDA-MB-231 (19,20). Although TMPRSS2 was induced by DEX in both the MCF-7 and MDA-MB-231 cells, MCF-7 was selected and used in the following experiments due to having a relatively low basal level of TMPRSS2. Certainly, some key findings that obtained from MCF-7 were further validated in MDA-MB-231, as shown in the supplementary figures. To establish the distribution of TMPRSS2 in the DEX-treated MCF-7 cells, we did indirect immunofluorescence staining with the specific anti-TMPRSS2 antibody. The intensity of the TMPRSS2 signal was remarkably increased 48 h following DEX treatment, particularly in the 1 µM DEX-treated cells (Figure 1C). We revealed that TMPRSS2 was predominantly localized in the cell peripheral membrane. In the cytoplasmic region, some vesicles of TMPRSS2 were also revealed (Figure 1C).
Increased migration is mediated by TMPRSS2 in DEX-treated breast cancer cells
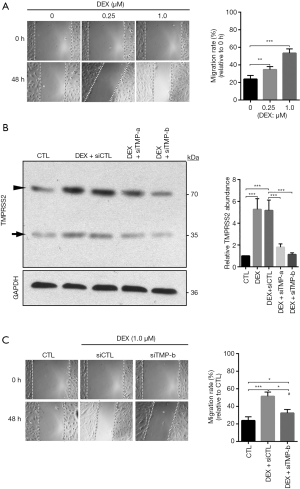
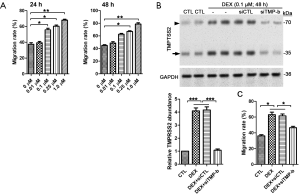
To explore the effects of DEX on the migration of breast cancer cells, scratch assay was carried out. We found that DEX increased the migration rate of MCF-7 and MDA-MB-231 in a dose-dependent manner (Figure 2A, Figure S2A). To test if TMPRSS2 involved DEX-induced increase in cellular migration, we knocked down TMPRSS2 expression with two different siRNAs specifically targeted against the human TMPRSS2 gene. In comparison with the control siRNAs, siTMP-a and especially siTMP-b, significantly decreased the protein levels of TMPRSS2 in DEX-treated MCF-7 (Figure 2B). A scratch assay was then carried out with DEX-treated MCF-7 that expressed siTMP-b. Our results showed that the increased migration rate was suppressed significantly in DEX-treated MCF-7 with TMPRSS2 knockdown (Figure 2C). The knockdown of TMPRSS2 was also discovered to significantly inhibit the cellular migration of DEX-treated MDA-MB-231 (Figure S2B,C). These findings indicate that DEX increases the migration of breast cancer cells in vitro by upregulating TMPRSS2.
Activation of the α2-adrenergic receptor/STAT3 signaling is responsible for the induction of TMPRSS2 expressions in DEX-treated MCF-7
DEX, an α2-adrenergic agonist, induces downstream signaling via the activation of α2-adrenergic receptors (2,3). There are three subtypes of Alpha 2-adrenergic receptor, and it has been shown each of them presents in MCF-7 cells (21). It was recently reported that alpha 2A subtype overexpression was associated with a better disease-free survival in patients with luminal tumors, while the overexpression of alpha 2C subtype was associated with worse prognosis for breast cancer patients with basal-like tumors (4,22). Nevertheless, the role of the alpha 2B subtype has been the subject of fewer studies on the pathogenesis of breast cancer. This study focused on the alpha 2B subtype by using the antibody ABN1481, which highly and specifically targets the alpha 2B subtype. In the current study, the expression of α2-adrenergic receptor was significantly increased at 48 h in 1 µM of DEX-treated MCF-7 (Figure 3A). Indirect immunofluorescence also revealed obvious colocalization of TMPRSS2 and α2-adrenergic receptors in the DEX-treated cells (Figure 3B). In addition, we found the levels of the phosphorylated STAT3Tyr705 in the nuclear fractions to be significantly increased at 24 and 48 h after DEX treatment (Figure 3C), suggesting that the STAT3 signaling pathway is activated. To investigate the role of the STAT3 signaling pathway in breast cancer, its specific inhibitor WP1006 was applied in this study. Our data showed that WP1006 at 5 and 10 µM dramatically inhibited DEX-induced elevation of the phospho-STAT3Tyr705 (Figure 3D). Moreover, the administrated WP1006 significantly suppressed DEX-induced upregulation of both full-length TMPRSS2 and its cleaved protease domain (Figure 3E). These results suggest that DEX upregulates the expression of TMPRSS2 possibly by activating the α2-adrenergic receptor/STAT3 signaling pathway.
DEX increases the secretion of the TMPRSS2 cleaved protease domain via Rab11 in breast cancer cells
The cleaved protease domain of TMPRSS2 has been found to be secreted into the cell media after autocleavage (14). We thus assessed the level of TMPRSS2 in the culture media from DEX-treated MCF-7 cells. The data from the ELISA assay showed that DEX induced a significant dose- and time- dependent increase of TMPRSS2 in the media (Figure 4A), evidencing that TMPRSS2 was secreted and present in the cultured media following treatment with DEX. We also isolated exosomes from the cultured media, and tested for the presence of TMPRSS2. Increases in exosomal markers such as a disintegrin and metalloproteinase domain-containing protein 10 (ADAM10) and heat shock protein 70 (Hsp70) were detected in the isolated exosomes by Western blot assay (Figure 4B), suggesting that the exosomes had been successfully isolated and obtained. In our study, Hsp90, another exosomal marker revealed no significant difference in the isolated exosomal fractions of the DEX-treated cells when compared with the controls. Hsp90 was therefore used as the loading control for the quantification of exosomal proteins (Figure 4B). As expected, a dose-dependent increase of the TMPRSS2 cleaved protease domain was significantly detected in the isolated exosomes from the cultured media of DEX-treated MCF-7 (Figure 4B).
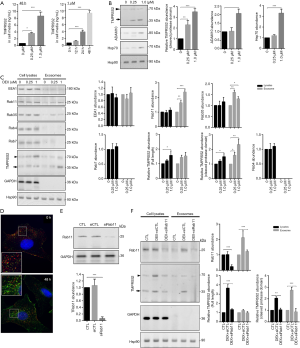
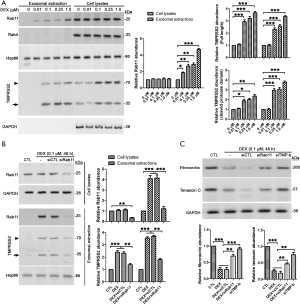
To reveal the mechanisms by which TMPRSS2 was secreted, we analyzed the levels of multiple vesicle-related markers, including the early endosome antigen (EEA1) and the late endosome (Rab7) markers as well as the recycling endosomal markers (Rab4 and Rab11) and the exosomal markers (Rab27 and Rab35). In the isolated exosomes, we detected a significant increase of Rab11 and Rab35, rather than EEA1, Rab4, and Rab7 (Figure 4C). Consistently, increases in full-length TMPRSS2 were mainly detected in total cell lysates, whereas increases of the TMPRSS2 cleaved protease domain were significantly detected in the exosomal fractions in DEX-treated MCF-7 cells (Figure 4C). The TMPRSS2 cleaved protease domain and Rab11 were also detected in DEX-treated MDA-MB-231 cells (Figure S3A). Rab11 acts in conjunction with a protein complex known as the exocyst to mediate terminal steps in cargo transport by recycling endosomes to cell-cell junctions (23). We thus performed indirect immunofluorescence to image the location of TMPRSS2 and Rab11. Obvious colocalization of TMPRSS2 and Rab11 was observed in the DEX-treated MCF-7 cells. This provides further evidence of the presence of TMPRSS2 in Rab11-positive exosomes (Figure 4D), indicating that Rab11 could play an important role in mediating the secretion of the TMPRSS2 cleaved protease domain. Concurrently, Rab11 was knocked down using the siRNAs specifically targeted to the two isoforms of human Rab11 (Rab11a and Rab11b) in cultured MCF-7 and MDA-MB-231 (Figure 4E, Figure S3B). DEX-induced increase of the TMPRSS2 cleaved protease domain in the exosomes was dramatically inhibited by siRab11 (Figure 4F, Figure S3B). These data suggest that Rab11 mediates DEX-induced secretion of the cleaved protease domain of TMPRSS2. In addition, increases of full-length TMPRSS2 and the TMPRSS2 cleaved protease domain in total cellular lysates were also significantly prevented by siRab11 (Figure 4F, Figure S3B), suggesting that overproduced TMPRSS2 may undergo degradation, most likely as a result of the blockage of the secretion pathway of TMPRSS2 caused by Rab11 deficiency.
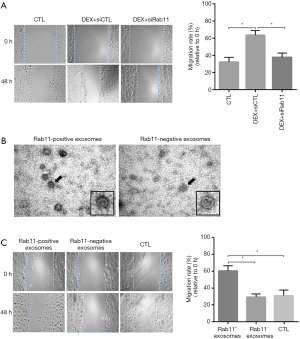
Rab11 increases the migration of DEX-treated human breast cancer cells
Our data demonstrated the mediating effect of Rab11 on the secretion of TMPRSS2 into exosomes following treatment with DEX treatment. To further assess the regulatory role of Rab11 and exosomal TMPRSS2 in the migration of DEX-treated MCF-7, we investigated the effects of Rab11 knockdown on the migration rate in DEX-treated MCF-7 cells. In comparison with the control siRNAs, the increase in the migration rate of the DEX-treated MCF-7 cells was remarkably reduced by siRab11 (Figure 5A). As described above, Rab11 was mainly present in the exosomes of the DEX-treated MCF-7 cells. To further elucidate the role of exosomal Rab11, we isolated Rab11-positive or negative exosomes from DEX-treated MCF-7 that expressed the control siRNA or siRab11, respectively, and then applied them to the other normally cultured MDA-MB-231 cell line. Firstly, we imaged the exosomes with a transmission electron microscope, finding that the Rab11-positive exosomes appeared to be much darker than the Rab11-negative exosomes (Figure 5B), which was indicative that more components in the exosomes were enriched by Rab11. Then, we investigated the effects of the Rab11-positive and negative exosomes on cellular migration in normally cultured MDA-MB-231. We found that compared to the non-treated cells, migration was significantly increased by the Rab11-positive exosomes, but not by the Rab11-negative exosomes (Figure 5C). These findings indicate that Rab11 that is present in exosomes play a critical role in the DEX-induced migration of breast cancer cell in vitro.
DEX promotes the ECM degradation through TMPRSS2 in the cultured breast cancer cells
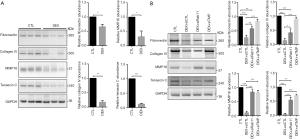
Degradation of the ECM promotes migration in various types of cells, such as prostate cancer cells (13). We assessed the alterations of several ECM proteins in DEX-treated MCF-7 and MDA-MB-231 cell lines. As expected, the protein levels of fibronectin, collagen IV, matrix metallopeptidase 16, and tenascin C were significantly downregulated by DEX (Figure 6A, Figure S3C). Moreover, the knockdown of Rab11 or TMPRSS2 remarkably inhibited DEX-induced downregulation of the ECM components (Figure 6B, Figure S3C). Therefore, our findings demonstrate that TMPRSS2 expression and secretion is elevated by DEX, thus inducing ECM degradation, which is responsible for the increase in breast cancer cell migration in vitro.
Discussion
DEX, a lipophilic α2 adrenergic agonist, has been widely used in surgery as an anesthetic to effectively reduce stress reaction and systemic inflammation and maintain the normal function of the cardiovascular system (16). TMPRSS2 has been found to be upregulated by androgenic hormones in prostate cancer cells and downregulated in androgen-independent prostate cancer tissue (14). In patients with prostate cancer, TMPRSS2 has been identified as a key driver of metastasis and a marker for poor prognosis (12-14). The combined effects of TTSP variants and their related genes, including TMPRSS2, were reported in relation to risk and patient outcome in breast cancer (10). Thus, we hypothesized that DEX may promote breast cancer cell migration and metastasis in vitro by regulating TMPRSS2 and its downstream signaling.
TMPRSS2 belongs to the serine protease family and contains a type II transmembrane domain, a receptor class A domain, a scavenger receptor cysteine-rich domain, and a protease domain (24). Serine proteases are known for their involvement in many physiological and pathological processes, and the protease domain of TMPRSS2 is thought to be cleaved and secreted into cell media after autocleavage (14). In this study, increases in both full-length TMPRSS2 and its cleaved protease domain were detected in two widely used breast cancer cell lines, MCF-7 and MDA-MB-231, following treatment with DEX. Consistently, it has been reported that the full-length TMPRSS2 and its cleaved protease domain were both present in cultured MCF-7, albeit with a lower level relative to head and neck cancer cells HSC-3 and SCC-9 as well as prostate cancer cells LNCaP (15). In this study, we also investigated the mechanism by which TMPRSS2 expression is induced by DEX. Firstly, we detected elevated expressions of α2-adrenergic receptors and the colocalization of α2-adrenergic receptors with TMPRSS2 along the cell membrane in DEX-treated MCF-7. The expression of TMPRSS2 has been reported to be significantly reduced by galiellalactone, a small-molecule STAT3 inhibitor, in both prostate and breast cancer cells (25). Moreover, galiellalactone has also been observed to inhibit the growth of triple-negative breast cancer cell lines (26). In our study, hosphor-STAT3 in the tyrosine 705 residue was detected to be elevated in the nuclear fractions of DEX-treated MCF-7. Interestingly, the inhibition of STAT3 by WP1006 significantly reduced DEX-mediated upregulation of TMPRSS2. Therefore, our data demonstrate that DEX can elevate the expression of TMPRSS2 partly through the activation of the α2-adrenergic receptor/STAT3 signaling pathway.
TMPRSS2 was detected in the supernatant media of the cultured HSC-3 cells by ELISA over a 72-h period (15), which suggests that TMPRSS2 can be released into the cancer microenvironment. Here, the elevated TMPRSS2 was also revealed by ELISA in the cultured media in DEX-treated MCF-7 cells, indicating that TMPRSS2 was produced, cleaved, and secreted into the media following DEX treatment. TMPRSS2 can induce proteolytic activity, resulting in the degradation of the components of the ECM (13). In this study, the degradation of ECM components such as fibronectin, collagen IV, matrix metallopeptidase 16, and tenascin C was detected in the DEX-treated MCF-7 and MDA-MB-231 cells, but not in the cells with knocked-down TMPRSS2. These findings suggest that TMPRSS2 may play an important role in DEX-induced migration of breast cancer cells, at least partly, through the degradation of the components of the ECM. Therefore, we assessed the migration rate of MCF-7 and MDA-MB-231 cells following DEX treatment. In our study, a dose-dependent increased migration rate was detected following DEX treatment over 24- and 48-h periods. As expected, the knockdown of TMPRSS2 significantly decreased DEX-induced migration, providing evidence that DEX can increase breast cancer cell migration by upregulating TMPRSS2. In the current study, a higher basal expression level of TMPRSS2 was detected in MDA-MB-231 than in MCF-7, which may be an important factor to explain why the migration rate of MDA-MB-231 is higher than MCF-7 under normal conditions (19,20).
We detected elevated TMPRSS2 in the cultured media of DEX-treated MCF-7, which is indicative of the presence of TMPRSS2 in the extracellular vesicles. Exosomes, one of the most notable populations of extracellular vesicle, are released by viable cells under normal conditions (27). TMPRSS2 has previously been detected in extracellular vesicles isolated from the prostate cancer cell LNCaP (28,29). Thus, we isolated the exosomes from the cultured media of DEX-treated MCF-7 and MDA-MB-231, and found a dramatic increase in the cleaved protease domain of TMPRSS2. To reveal the potential pathway responsible for TMPRSS2 secretion, we analyzed the levels of several extracellular vesical markers in the isolated exosomes. A significant increase of Rab11 was detected in relation to Rab35, whereas the early endosomal marker EEA1, the recycling endosomal marker Rab4, and the late endosomal marker Rab7 were not detected in the isolated exosomes. In addition, the colocalization of TMPRSS2 and Rab11 was clearly revealed in the DEX-treated MCF-7 cells. Similarly, Rab11 has also been identified in exosomes isolated from prostate cancer cell line PC346 (29). Rab11, a member of the Rab family within the GTPase superfamily, is associated with both the constitutive and regulated secretory pathways, and may be involved in protein transport (30). Here, the two isoforms of Rab11, Rab11a and Rab11b, were simultaneously knocked down using the siRNAs specifically targeted against human Rab11a and Rab11b. The Rab11 knockdown reduced the abundance of the cleaved protease domain of TMPRSS2 in the exosomal fractions isolated from the DEX-treated MCF-7 and MDA-MB-231 cells, which indicates that Rab11 mediates the secretion of cleaved TMPRSS2 into the culture media of breast cancer cells following DEX treatment in vitro. In mitotic cells, the localization of TMPRSS2 to recycling endosomes in interphase and their transport to the intercellular bridge during cytokinesis depend on Rab11 (31). These findings demonstrate the crucial role Rab11 plays in the regulation of endocytic and recycling events. Moreover, the DEX-induced degradation of the ECM components was remarkably prevented by the knockdown of Rab11 in the two breast cancer cell lines. It is further supported by the observation that compared to Rab11-negative exosomes, the application of Rab11-positive exosomes isolated from DEX-treated MCF-7 significantly increased migration in normally cultured MDA-MB-231. Moreover, transmission electronic microscopy shows that the components of the Rab11-positive exosomes were likely to be much richer than those of the Rab11-negative exosomes. The role of exosomes in the migration of breast cancer cells should be further investigated.
Conclusions
We demonstrated that DEX increases migration in breast cancer cells in vitro through TMPRSS2-mediated degradation of ECM components. We also showed that DEX upregulates the expression of TMPRSS2 through the activation of the α2-adrenergic receptor/STAT3 signaling pathway, and increases the secretion of the cleaved protease domain of TMPRSS2 through exosomal Rab11. Our findings provide in vitro evidence of the deleterious effects of DEX. More in vivo research to assess the influence of the perioperative DEX application on the outcomes of breast cancer, particularly studies of a clinical nature, is required.
Acknowledgments
Funding: This study was supported by the National Nature Science Foundation of China (grant numbers 81771198, 2017) and Haiyan Foundation of Harbin Medical University Cancer Hospital (grant number HY-QNJJ2018039).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.04.28). Dr. MC reports grants from the National Nature Science Foundation of China, grants from Haiyan Foundation of Harbin Medical University Cancer Hospital, during the conduct of the study; Dr. XS reports grants from the National Nature Science Foundation of China, grants from Haiyan Foundation of Harbin Medical University Cancer Hospital, during the conduct of the study; Dr. XH reports grants from the National Nature Science Foundation of China, grants from Haiyan Foundation of Harbin Medical University Cancer Hospital, during the conduct of the study; Dr. XW reports grants from the National Nature Science Foundation of China, grants from Haiyan Foundation of Harbin Medical University Cancer Hospital, during the conduct of the study; Dr. PZ reports grants from the National Nature Science Foundation of China, grants from Haiyan Foundation of Harbin Medical University Cancer Hospital, during the conduct of the study; Dr. GW reports grants from the National Nature Science Foundation of China, grants from Haiyan Foundation of Harbin Medical University Cancer Hospital, during the conduct of the study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Mille R KD. JeMal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Hwang JH, Chang IS, Park SW, et al. Sole use of dexmedetomidine for sedation and analgesia in patients undergoing endovenous thermal ablation for incompetent saphenous veins. Ann Transl Med 2019;7:638. [Crossref] [PubMed]
- Fairbanks CA, Stone LS, Wilcox GL. Pharmacological profiles of alpha 2 adrenergic receptor agonists identified using genetically altered mice and isobolographic analysis. Pharmacol Ther 2009;123:224-38. [Crossref] [PubMed]
- Powe DG, Voss MJ, Habashy HO, et al. Alpha- and beta-adrenergic receptor (AR) protein expression is associated with poor clinical outcome in breast cancer: an immunohistochemical study. Breast Cancer Res Treat 2011;130:457-63. [Crossref] [PubMed]
- Shkurnikov MY, Galatenko VV, Lebedev AE, et al. On statistical relationship between ADRA2A expression and the risk of breast cancer relapse. Bull Exp Biol Med 2014;157:454-58. [Crossref] [PubMed]
- Xia M, Ji NN, Duan ML, et al. Dexmedetomidine regulate the malignancy of breast cancer cells by activating α2-adrenoceptor/ERK signaling pathway. Eur Rev Med Pharmacol Sci 2016;20:3500-6. [PubMed]
- Lavon H, Matzner P, Benbenishty A, et al. Dexmedetomidine promotes metastasis in rodent models of breast, lung, and colon cancers. Br J Anaesth 2018;120:188-96. [Crossref] [PubMed]
- Liu Y, Sun J, Wu T, et al. Effects of serum from breast cancer surgery patients receiving perioperative dexmedetomidine on breast cancer cell malignancy: A prospective randomized controlled trial. Cancer Med. 2019;8:7603-12. [Crossref] [PubMed]
- Bugge TH, Antalis TM, Wu Q. Type II transmembrane serine proteases. J Biol Chem 2009;284:23177-81. [Crossref] [PubMed]
- Luostari K, Hartikainen JM, Tengström M, et al. Type II transmembrane serine protease gene variants associate with breast cancer. PLoS One 2014;9:e102519. [Crossref] [PubMed]
- Matulay JT, Wenske S. Genetic signatures on prostate biopsy: clinical implications. Transl Cancer Res 2018;7:S640-50. [Crossref]
- Chen YW, Lee MS, Lucht A, et al. TMPRSS2, a serine protease expressed in the prostate on the apical surface of luminal epithelial cells and released into semen in prostasomes, is misregulated in prostate cancer cells. Am J Pathol 2010;176:2986-96. [Crossref] [PubMed]
- Ko CJ, Huang CC, Lin HY, et al. Androgen-Induced TMPRSS2 Activates Matriptase and Promotes Extracellular Matrix Degradation, Prostate Cancer Cell Invasion, Tumor Growth, and Metastasis. Cancer Res 2015;75:2949-60. [Crossref] [PubMed]
- Afar DE, Vivanco I, Hubert RS, et al. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res 2001;61:1686-92. [PubMed]
- Lam DK, Dang D, Flynn AN, et al. TMPRSS2, a novel membrane-anchored mediator in cancer pain. Pain 2015;156:923-30. [Crossref] [PubMed]
- Weerink MAS, Struys MMRF, Hannivoort LN, et al. Clinical Pharmacokinetics and Pharmacodynamics of Dexmedetomidine. Clin Pharmacokinet 2017;56:893-913. [Crossref] [PubMed]
- Ebert TJ, Hall JE, Barney JA, et al. The effects of increasing plasma concentrations of dexmedetomidine in humans. Anesthesiology 2000;93:382-94. [Crossref] [PubMed]
- Fujita Y, Inoue K, Sakamoto T, et al. A comparison between dosages and plasma concentrations of dexmedetomidine in clinically ill patients: a prospective, observational, cohort study in Japan. J Intensive Care 2013;1:15. [Crossref] [PubMed]
- Hooshmand S, Ghaderi A, Yusoff K, et al. Downregulation of RhoGDIα increased migration and invasion of ER (+) MCF7 and ER (-) MDA-MB-231 breast cancer cells. Cell Adh Migr 2013;7:297-303. [Crossref] [PubMed]
- Razak NA, Abu N, Ho WY, et al. Cytotoxicity of eupatorin in MCF-7 and MDA-MB-231 human breast cancer cells via cell cycle arrest, anti-angiogenesis and induction of apoptosis. Sci Rep 2019;9:1514. [Crossref] [PubMed]
- Castillo LF, Rivero EM, Goffin V, et al. Alpha2-adrenoceptor agonists trigger prolactin signaling in breast cancer cells. Cell Signal 2017;34:76-85. [Crossref] [PubMed]
- Rivero EM, Martinez LM, Bruque CD, et al. Prognostic significance of α- and β2-adrenoceptor gene expression in breast cancer patients. Br J Clin Pharmacol 2019;85:2143-54. [Crossref] [PubMed]
- Guichard A, Nizet V, Bier E. RAB11-mediated trafficking in host-pathogen interactions. Nat Rev Microbiol 2014;12:624-34. [Crossref] [PubMed]
- Szabo R, Wu Q, Dickson RB, et al. Type II transmembrane serine proteases. Thromb Haemost 2003;90:185-93. [Crossref] [PubMed]
- Handle F, Puhr M, Schaefer G, et al. The STAT3 Inhibitor Galiellalactone Reduces IL6-Mediated AR Activity in Benign and Malignant Prostate Models. Mol Cancer Ther 2018;17:2722-31. [Crossref] [PubMed]
- Kim HS, Kim T, Ko H. Identification of galiellalactone-based novel STAT3-selective inhibitors with cytotoxic activities against triple-negative breast cancer cell lines. Bioorg Med Chem 2017;25:5032-40. [Crossref] [PubMed]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200:373-83. [Crossref] [PubMed]
- Lázaro-Ibáñez E, Lunavat TR, Jang SC, et al. Distinct prostate cancer-related mRNA cargo in extracellular vesicle subsets from prostate cell lines. BMC Cancer 2017;17:92. [Crossref] [PubMed]
- Jansen FH, Krijgsveld J, van Rijswijk A, et al. Exosomal secretion of cytoplasmic prostate cancer xenograft-derived proteins. Mol Cell Proteomics 2009;8:1192-205. [Crossref] [PubMed]
- Welz T, Wellbourne-Wood J, Kerkhoff E. Orchestration of cell surface proteins by Rab11. Trends Cell Biol 2014;24:407-15. [Crossref] [PubMed]
- Takahashi S, Takei T, Koga H, et al. Distinct roles of Rab11 and Arf6 in the regulation of Rab11-FIP3/arfophilin-1 localization in mitotic cells. Genes Cells 2011;16:938-50. [Crossref] [PubMed]


