
Nosocomial cross-infection of hypervirulent Listeria monocytogenes sequence type 87 in China
Introduction
Listeria monocytogenes is a pathogen of animals and humans, which can be transmitted through food and water. Well-defined population groups, such as the elderly, immune compromised, the immunocompromised pregnant women and neonates, are particularly susceptible to listeriosis, and the average mortality rate is 20% to 35% (1-3). Human listeriosis is a major public health challenge globally (4) and outbreaks associated with L. monocytogenes are increasing (1,5,6).
L. monocytogenes can be divided into four phylogenetic lineages, four PCR sero-groups and 13 serotypes. Serotypes 1/2a (lineage II), is predominantly associated with food-derived sources, along with serotypes 1/2b and 4b (lineage I) are responsible for 95% of human cases reported (7). Detection of these different serotypes can help us to determine its geographical origin; for example, serotypes 1/2a and 4b are prevalent in some European countries, e.g., Finland (8), Sweden (9), Italy (10) and Hungary (11), and 4b is the predominant type in Japan (12) and US (13).
In China, the most prevalent serotype is serotype 1/2b accounting for more than half of all clinical isolates (64.29%), and serotypes 1/2a and 4b are ranked the second (21.43%) and third (7.14%), respectively (14). Feng et al. analyzed listeriosis cases from 2000 to 2009, and reported that the average mortality rate was 21%, but the mortality rate for newborns reached 56% (15). As L. monocytogenes is transmitted mainly through contaminated food, nosocomial cross-infection in neonates is rare but serious (16,17). In this study, we report a nosocomial cross-infection of L. monocytogenes sequence type (ST) 87 between two neonates.
On July 28, 2016, two cases of L. monocytogenes infection were identified in neonates through the Food borne Disease Monitoring and Reporting System of China. We culture the L. monocytogenes from the blood sample of the first neonate (named SD1) and the cerebrospinal fluid (CSF) sample of the second neonate (named SD3). Both of these isolates cultured from the neonates were serotype 1/2b, with an indistinguishable pulsotype, and were susceptible to a panel of antimicrobial compounds (Figure S1). Whole-genome sequencing (WGS) confirmed that the two isolates were ST87 with no detectable differences. These two isolates were positive for the Listeria pathogenicity island 4 (LIPI-4). Epidemiological investigation showed insufficient surface cleaning and lack of patient isolation measures may cause the risk factors of cross-contamination in this case. There searchers of epidemiological survey confirmed the fact that both neonates were nursed by the same group of nurses and without strict sterilization.

Methods
Medical ethics
A signed informed consent was obtained from the families of the children, and the research plan was approved by the Medical Ethics Committee of the Shandong Center for Disease Control and Prevention (CDC, Jinan, China).
Epidemiological and environmental investigations
On July 29th, 2016, we interviewed the maternity staff in Hospital A (in Jinan city) to collect clinical, diagnostic laboratory, demographic and epidemiological data, including the epidemiological questionnaires used in a survey on potential food exposures program during the 28 days before illness onset. On August 2nd, 2016, according to the case information and pulse-field gel electrophoresis (PFGE) results (Figure S1), the investigating team went to Hospital B (in Shanghe City) where both of these children born. Through field investigation and discussions with clinicians and nurses, we got all the details of the diagnostic and therapeutic measures in Hospital B. We collected anal swabs from the two parturients in Hospital A, along with 11 environmental swab samples and 2 food samples from the neonatal unit of Hospital B.
Microbial isolation and serotyping
All the samples collected during the investigation were sent to Shandong Centers for Disease Control and Prevent laboratory for microbiological culture. Serotypes were determined using the multiplex PCR assays, followed by the antigen serum agglutination tests (18).
Antimicrobial susceptibility testing
Antimicrobial susceptibility was tested by the broth microdilution method according to the standard protocols of the Clinical & Laboratory Standards Institute (CLSI) (19) and European committee for the testing of antimicrobial susceptibility (EUCAST) (20). A panel of six antimicrobial compounds was used, including ampicillin (AMP), penicillin, tetracycline (TET), meropenem, trimethoprim-sulfamethoxazole and erythromycin. Staphylococcus aureus ATCC™29213 was used as a quality control bacterium. Minimal inhibitory concentrations (MICs) were interpreted according to CLSI and EUCAST guidelines.
Pulse field gel electrophoresis (PFGE)
The PFGE of the isolates was performed using the PulseNet standardized protocol (21). Restriction endonuclease digestion was carried out using ApaI and AscI, respectively (Takara, Dalian, China), and run on a CHEF-DR III (Bio-Rad Laboratories, Hercules, CA, USA) over 19 h on 1% SeaKem gold agarose (Lonza, Rockland, MF) in 0.5 X Tris-borate-EDTA. BioNumerics software (Applied Maths, Kortrijk, Belgium) was used to calculate the percent similarity (Dice coefficient) of PFGE banding patterns as we described before (22).
WGS
Genomic DNA for all the isolates was harvested using the wizard genomic DNA purification kit (Promega, USA). DNA libraries were prepared using the KAPA Low-Throughput Library Preparation Kit with Standard PCR Amplification Module (KapaBiosystems, Wilmington, USA), following the manufacturer’s instructions except for the following modifications: 750 ng DNA was sheared using an M220 instrument (Covaris, Woburn, USA) in a 50 μL screw cap microtube at 50 peak power, 20 duty factor, 20 °C, 200 cycles per burst and 25s duration. Adapter-ligated fragments were size-selected to 700–800 bp following Illumina protocols. Standard desalted TruSeq LT and PCR primers were obtained from Integrated DNA Technologies (Coralville, IA) and used at 0.375 and 0.5 µM final concentrations, respectively. The PCR assay was reduced to 4 cycles. Libraries were quantified using the KAPA Library Quantification Kit (Kapa Biosystems, Wilmington, USA), with a 10 µL volume and 90s annealing/extension PCR (the other steps are the same as the manufacturer’s protocols), then pooled and normalized to 4 nM. Pooled libraries were re-quantified by ddPCR on a QX200 system (Bio-Rad, USA), using the Illumina TruSeq ddPCR Library Quantification Kit following the manufacturer’s protocols, except for an extended 2 min annealing/extension time. The libraries were sequenced 2×150 nucleotide paired-end v2 on the HiSeq platform (Illumina, USA) at 13.5 pM, following the manufacturer’s protocols.
Genome assembly and annotation
Raw read quality was assessed with Fast QC (v0.11.5) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and low-quality sequences were trimmed using Trimmomatic (v0.36) (23). The high-quality read data was assembled into contigs using SPAdes v3.9.1 (24). Gene prediction and annotation were performed using Prokka version 1.12-beta (25). Phage sequences were screened using PHAST (26).
In silico sub-typing by core genome multilocus sequence typing (cgMLST), multilocus sequence typing (MLST) and population analysis
The cgMLST scheme consists of 1,748 highly conserved core genes from the L. monocytogenes EGD-e reference strain. This genotyping method defines cgMLST types (CTs) as groups of cgMLST profiles that differ by up to 7 allelic mismatches and sublineage (SL) as groups of cgMLST profiles that differ by up to 150 allelic mismatches out of 1,748 loci. The cgMLST analysis was performed using the BIGSdb-Lm platform bioinformatics tool (http://bigsdb.pasteur.fr/listeria) (27,28). To elucidate the relationship of our isolates with the prevalent subtypes, published genome assemblies of L. monocytogenes were downloaded from GenBank database (accessed on March 21st, 2018). We used the 7-gene MLST scheme from the BIGSdb-Lm to perform subtyping in silico by BLAST for all the genomes (27). Minimum spanning trees of the two dominant lineages, lineages I and II, were generated using the eBurst algorithm in PHYLOViZ software (29).
Bioinformatic analyses of WGS data
Assembled genomes were screened for the absence/presence of antimicrobial resistance genes (AMR) using the ABRicate (https://github.com/tseemann/abricate) software package, which contains the ResFinder 2.1 database (30). Biocide resistance genes (tetR, tnpABC, qacH, bcrABC, emrE, emrC and qacC) and comK were screened for using the BLASTN algorithm with a minimum nucleotide identity and alignment length coverage of 80% (31).
Assessment of virulence factors
The presence and integrity of virulence factors was assessed using L. monocytogenes EGD-e (accession no: NC_003210) as the reference genome for internalin A (inlA), internalin B (inlB), Listeria pathogenicity island 1 (LIPI-1) and stress survival islet 1 (SSI-1). L. monocytogenes F2365 was used as reference genome for listeria pathogenicity island 3 (LIPI-3) with the protein sequences LMOf2365_1113 to LMOf2365_1119, whilst L. monocytogenes LM9005581 was used as reference for LIPI-4 with the protein sequences (LM9005581_70009 to LM9005581_70014). Analysis was performed using the BLASTP algorithm with a minimum amino acid identity of 70%, allowing the identification of premature stop codons and internal deletions.
Single-nucleotide polymorphisms analysis
Chromosomal SNPs calling analysis was performed using MUMmer v3.23 with the default parameter (32). SNP analyses were performed separately in comparison to the chromosomal sequence from the reference ST87 strain isolated from a patient with septicemia in Beijing, China (accession no: CP015593). SNP matrices were generated based on results of each comparison and then used to construct phylogenetic tree using maximum likelihood method via MEGA v5.1 (33), with bootstrap value set to 1000 (34).
Results
Neonate cases and related epidemiological features
On July 14th and 21st, 2016, two neonate cases were reported by Hospital A. The timeline of the events is shown in Figure 1. On July 9th, 2016; neonate-1 presenting with pneumonia, bacteremia, hypoxic ischemic encephalopathy (HIE) and low-grade asphyxiation was admitted to Hospital A. One L. monocytogenes isolate (SD1) was obtained from blood culture on July 14th, 2016, at Hospital A. On July 17th, 2016, neonate-2 was admitted in Hospital A for suppurative meningitis, pneumonia, hyperbilirubinemia and asphyxiation. Another L. monocytogenes isolate (SD3) was also isolated from a CSF culture on July 21st, 2016, in Hospital A.
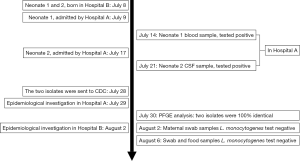
Interviews were conducted with the parturients on July 29th, 2016, and it was found that the two families lived in different geographical areas. The two pregnant women were healthy during pregnancy, especially for four antenatal weeks. The sole intersection of neonates-1 and neonates-2 was that they were born in the same hospital (Hospital B).
Investigations in Hospital B revealed that the two parturients were admitted to different wards across two rooms, did not meet before, during, or after birth, nor were the same medical and nursing staff present at the births. Neonate-1 was born by emergency caesarean section due to fetal distress at 37 weeks of gestation, and the amniotic fluid was contaminated (III level). Neonate-1 was resuscitated in the delivery room (APGAR scores of 5, 8 and 9 at 1, 5 and 10 min respectively) and diagnosed with neonatal sepsis, neonatal asphyxia, HIE and alimentary tract hemorrhage. Neonate-2 was born by natural labor at 35 weeks of gestation and healthy at birth (APGAR score of 10 at 1 min and clear amniotic fluid), but was diagnosed with premature birth, aspiration pneumonia and low birth weight. Then, both neonates were admitted to neonatal department of Hospital B through different channels within half an hour by different nurses, where they were weighed and placed in different but adjacent incubators. Neonate-1 was supported in a neonatal incubator near the incubator of neonate-2. The two neonates were cared for by the same group of nurses on the evening of July 9th, 2016. It is worth noting that there was no record of sanitation of the neonatal incubator number 64 on July 8–9th, 2016. No further cases were identified by the retrospective studies of medical records and following surveillance in Hospital B to date.
Sample culture and diagnosis
The anal swabs from the two parturients in Hospital A cultured were negative for L. monocytogenes. Three environmental samples from Hospital B were positive for S. haemolyticus (two samples) and S. gallinarum (one sample), which were not detected in the samples from both neonates (Table 1). Therefore, L. monocytogenes was considered the only causative agent for the two cases. Combined with the clinical symptoms and laboratory results, the two cases were diagnosed as listeriosis, despite the anal swabs culture from the two parturients were negative after the use of antibiotics.
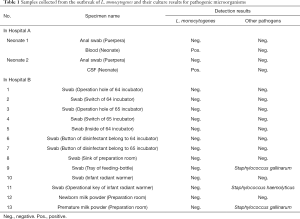
Full table
Molecular subtyping and phylogenetic relationship of L. monocytogenes isolates
The cgMLST profile of 1,748 loci was determined for the two L. monocytogenes isolates. They shared the same cgMLST profile (ST87). Seven housekeeping genes (abcZ, blgA, cat, dapE, dat, ldh, and lhkA) for MLST were extracted from WGS data showing that the ST of two isolates is ST87. Population study with the known STs showed that ST87 belonged to lineage I and the clonal complex (CC) 5 was the nearest large cluster (Figure S2).

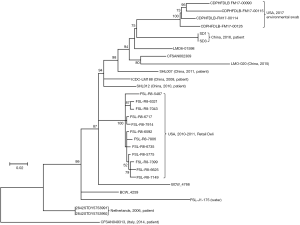
Comparative genomic analysis showed that the two isolates were indistinguishable, with no SNP differences being detected. Our data confirmed that the bacterial etiological agent identified in the two cases was considered to be the same, or the isolate was transmitted from neonate-1 to neonate-2. The phylogenetic analysis based on the whole genome SNPs of all published ST87 genomes showed that these two isolates, SD1 and SD3, were close to the isolates from patients in China reported previously. Further, all six Chinese ST87 isolates were dispersed in one large cluster along with several environmental isolates from the US and some other isolates without metadata, and three isolates from European patients were also clustered together distant from the branch containing the isolates from China. It is interesting that four isolates collected from the US in 2017 were closely related to the isolates in this study (Figure 2).
Resistance and virulence factors
According to antimicrobial resistance gene and stress island analysis, no antibiotic resistance genes were identified in any of the isolates sequenced when the genomes were queried against current databases, which is in line with the antimicrobial susceptibility test. Antibiotic resistance gene analysis suggested that there were no benzalkonium chloride (BC) tolerance genes qacH (encoded in Tn6188 transposon) and emrC (an efflux pump).
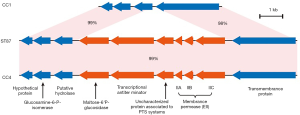
Regarding to the virulence genes, SSI-1 (encodes the genes denoted as lmo0444-lmo0448) was confirmed to be present in the isolates (Table S1). The presence and integration of the following virulence factors was also investigated: prfA, plcA, hly, actA (all located in LIPI-1), inlA, inlB, llsGHXBYDP (located in LIPI-3) and LM9005581_70009 to LM9005581_70014 (located in LIPI-4). LIPI-1, inlA, inlB and LIPI-4 were present in the isolates without of LIPI-3 (Table S1). The PTS operon of ST87 was highly similar to that of prevalent strain CC4 (Figure 3). Therefore, the ST87 isolates SD1 and SD3 were considered as hypervirulent strains.

Full table
Discussion
Although L. monocytogenes is a rare cause of nosocomial infection, there were a few reports of L. monocytogenes in neonates with neonatal sepsis in Israel, 2002 (17) and France, 2013 (16). L. monocytogenes cross-transmission via health workers has been suspected, but the mechanism of transmission remains to be established. To our knowledge, there has been no nosocomial cross-infection of L. monocytogenes reported in China. Here, we describe a nosocomial cross-infection of listeriosis.
Combining all the evidence (field investigation and laboratory analysis, the epidemiological, microbiological and genomic data), we can conclusively demonstrated that insufficient surface cleaning and unsatisfactory hand hygiene by the same group of nurses on the evening of July 9th, 2016, at the neonatal department during the time when these neonates were being cared for were the possible causes of cross-infection that potentially resulted in a nosocomial outbreak of hypervirulent L. monocytogenes ST87, which carried the recently identified PTS-LIPI-4.
To date, ST87 (serotype 1/2b) which represents a rare ST from lineage I is relatively uncommon in Western countries; only two outbreaks in Spain were reported (35). However, it is the predominant type in human listeriosis cases (14,36) and wild rodents from natural environments in China (37). Population study with known STs showed that ST87was closely related to the prevalent strain CC5 (Figure S2), a clonal group of serotypes 1/2b and 3b contaminating a variety of food products and processing environments and/or causing outbreaks (38,39). Analysis of L. monocytogenes isolated from food sources noted that ST87 (9.2%) was the third most prevalent ST in China (37); meanwhile, the first and second most common STs were ST9 (29.1%) and ST8 (10.7%), respectively (14. The high prevalence of ST87 in China, which is different from that found in the Western world, may be attributed to the different food matrices and dietary habits. Compared with all published ST87 genomes from different regions worldwide, all six isolates from patients in China exhibited large genome diversities with each other and with the exception of SD1 and SD3 from this study, did not cluster closely, suggesting that there was no dominant strain of ST87 contain SD1 and SD3 discovered in China until now. Therefore, further studies on epidemiological characteristics and pathogenic mechanism of strain ST87 contain SD1 and SD3 are required.
LIPI-4 is a recently identified gene cluster that expresses a PTS system. PTS contributes to the neural and placental infections of CC4 (40). Wang et al. analyzed 8 clinical strains of ST87, which were responsible for maternal-neonatal or central nervous system infections, harbored LIPI-4 gene (41). Our research showed that the operon encoding PTS system was positive in all previously published ST87 genomes, along with our two isolates. In addition to the LIPI-4 cluster, other virulence genes identified included those found in LIPI-1, inlA and inlB may provide further support for the observations of ST87 in listeriosis cases in China.
Antimicrobial resistance in bacterial pathogens has become an international public health issue. L. monocytogenes is no exception, and there were some reports of resistance to TET, AMP, cephalothin, streptomycin and penicillin G in L. monocytogenes isolates (42,43). However, the isolates from our cases, reported here, were susceptible to all the tested drugs. Genomic analysis for markers of antimicrobial resistance confirmed the observed phenotypes. This observation supports the empirical use of meropenem which is the most common drug of choice for treating encephalitis. No new cases were found in Hospital B, after the neonatal incubator number 64 (used by neonate-1) was disinfected. Despite these careful investigations no source was identified, especially for the first baby (neonate-1) who was infected from birth. It is most likely that pregnant women ate food contaminated with L. monocytogenes, leading to secondary intrauterine infections in newborns, so the government should strengthen food risk safety surveillance.
Conclusions
In summary, a nosocomial cross-infection of the virulent L. monocytogenes ST87 was investigated in China. Results of PFGE analysis and WGS analysis suggested that the two completely same L. monocytogenes strain was isolated in the two babies, and these isolates are considered to be hypervirulent due to the carriage of virulence factors including LIPI-1, the internalin genes (inlA, inlB) and LIPI-4. Considering the genomic data together with epidemiological evidence and clinical symptoms, inadequate surface cleaning and hand hygiene at the neonatal department of Hospital B was hypothesized as a possible cause of cross-infection. The test results of environmental samples in Hospital B indicate that strict sterilization and patient isolation measures cannot be emphasized enough in neonatal nursing.
Acknowledgments
Funding: This study was supported by Shandong Province Science Foundation for Talent Cultivation [ZR2015PH059]; Projects of medical and health technology development program in Shandong province [2016WS0383, 2018WS304]; the outstanding young technical backbone training project of Jinan [SFW]. The State Major Infectious Disease Research Program [2012ZX10004206-007, 2017zx10103004-007]; National Science and Technology Major Project of China [2018ZX10733402].
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Zhi-De Hu, Bing Gu) for the series “Advances in laboratory tests for infectious diseases” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-19-2743). The series “Advances in Laboratory Tests for Infectious Diseases” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study obtained signed informed consent from the families of children, and the research plan was approved by the Medical Ethics Committee of the Shandong Center for Disease Control and Prevention.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Allerberger F, Wagner M. Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect 2010;16:16-23. [Crossref] [PubMed]
- Madjunkov M, Chaudhry S, Ito S. Listeriosis during pregnancy. Arch Gynecol Obstet 2017;296:143-52. [Crossref] [PubMed]
- Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect 2007;9:1236-43. [Crossref] [PubMed]
- Chenal-Francisque V, Lopez J, Cantinelli T, et al. Worldwide distribution of major clones of Listeria monocytogenes. Emerg Infect Dis 2011;17:1110-2. [Crossref] [PubMed]
- Goulet V, Hedberg C, Le Monnier A, et al. Increasing incidence of listeriosis in France and other European countries. Emerg Infect Dis 2008;14:734-40. [Crossref] [PubMed]
- Laksanalamai P, Joseph LA, Silk BJ, et al. Genomic characterization of Listeria monocytogenes strains involved in a multistate listeriosis outbreak associated with cantaloupe in US. PLoS One 2012;7:e42448. [Crossref] [PubMed]
- McLauchlin J, Mitchell RT, Smerdon WJ, et al. Listeria monocytogenes and listeriosis: a review of hazard characterisation for use in microbiological risk assessment of foods. Int J Food Microbiol 2004;92:15-33. [Crossref] [PubMed]
- Lukinmaa S, Aarnisalo K, Suihko ML, et al. Diversity of Listeria monocytogenes isolates of human and food origin studied by serotyping, automated ribotyping and pulsed-field gel electrophoresis. Clin Microbiol Infect 2004;10:562-8. [Crossref] [PubMed]
- Parihar VS, Lopez-Valladares G, Danielsson-Tham ML, et al. Characterization of human invasive isolates of Listeria monocytogenes in Sweden 1986-2007. Foodborne Pathog Dis 2008;5:755-61. [Crossref] [PubMed]
- Mammina C, Aleo A, Romani C, et al. Characterization of Listeria monocytogenes isolates from human listeriosis cases in Italy. J Clin Microbiol 2009;47:2925-30. [Crossref] [PubMed]
- Kiss R, Tirczka T, Szita G, et al. Listeria monocytogenes food monitoring data and incidence of human listeriosis in Hungary, 2004. Int J Food Microbiol 2006;112:71-4. [Crossref] [PubMed]
- Miya S, Takahashi H, Nakagawa M, et al. Genetic characteristics of Japanese clinical Listeria monocytogenes isolates. PLoS One 2015;10:e0122902. [Crossref] [PubMed]
- Burall LS, Grim CJ, Datta AR. A clade of Listeria monocytogenes serotype 4b variant strains linked to recent listeriosis outbreaks associated with produce from a defined geographic region in the US. PLoS One 2017;12:e0176912. [Crossref] [PubMed]
- Wang Y, Jiao Y, Lan R, et al. Characterization of Listeria monocytogenes isolated from human Listeriosis cases in China. Emerg Microbes Infect 2015;4:e50. [Crossref] [PubMed]
- FengYanfang RL. Zhang Lishi. Lisisteriosis case reported in medical literatures in China, 2000-2009. Dis Surveill 2011;26:6.
- Lazarus C, Leclercq A, Lecuit M, et al. Probable nosocomial transmission of listeriosis in neonates. J Hosp Infect 2013;85:159-60. [Crossref] [PubMed]
- Colodner R, Sakran W, Miron D, et al. Listeria monocytogenes cross-contamination in a nursery Am J Infect Control 2003;31:322-4. [corrected]. [Crossref] [PubMed]
- Doumith M, Buchrieser C, Glaser P, et al. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 2004;42:3819-22. [Crossref] [PubMed]
- M45 Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated of Fastidious Bacterial. Available online: https://clsi.org/media/1450/m45ed3_sample.pdf
- Testing ECoAS. European antimicrobial breakpoints. Basel: EUCAST, 2017.
- Ribot EM, Fair MA, Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 2006;3:59-67. [Crossref] [PubMed]
- Li XP, Gao RH, Hou PB, et al. Characterization of the Salmonella enterica Serotype Isangi Isolated from Patients for the First Time in China. Foodborne Pathog Dis 2017;14:427-31. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19:455-77. [Crossref] [PubMed]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068-9. [Crossref] [PubMed]
- Zhou Y, Liang Y, Lynch KH, et al. PHAST: a fast phage search tool. Nucleic Acids Res 2011;39:W347-52. [Crossref] [PubMed]
- Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010;11:595. [Crossref] [PubMed]
- Moura A, Criscuolo A, Pouseele H, et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol 2016;2:16185. [Crossref] [PubMed]
- Nascimento M, Sousa A, Ramirez M, et al. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2017;33:128-9. [Crossref] [PubMed]
- Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012;67:2640-4. [Crossref] [PubMed]
- Müller A, Rychli K, Muhterem-Uyar M, et al. Tn6188 - a novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLoS One 2013;8:e76835. [Crossref] [PubMed]
- Delcher AL, Phillippy A, Carlton J, et al. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 2002;30:2478-83. [Crossref] [PubMed]
- Kumar S, Nei M, Dudley J, et al. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 2008;9:299-306. [Crossref] [PubMed]
- Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 2010;5:e9490. [Crossref] [PubMed]
- Pérez-Trallero E, Zigorraga C, Artieda J, et al. Two outbreaks of Listeria monocytogenes infection, Northern Spain. Emerg Infect Dis 2014;20:2155-7. [Crossref] [PubMed]
- Huang YT, Ko WC, Chan YJ, et al. Disease Burden of Invasive Listeriosis and Molecular Characterization of Clinical Isolates in Taiwan, 2000-2013. PLoS One 2015;10:e0141241. [Crossref] [PubMed]
- Wang Y, Lu L, Lan R, et al. Isolation and characterization of Listeria species from rodents in natural environments in China. Emerg Microbes Infect 2017;6:e44. [Crossref] [PubMed]
- Chen Y, Gonzalez-Escalona N, Hammack TS, et al. Core Genome Multilocus Sequence Typing for Identification of Globally Distributed Clonal Groups and Differentiation of Outbreak Strains of Listeria monocytogenes. Appl Environ Microbiol 2016;82:6258-72. [Crossref] [PubMed]
- McCollum JT, Cronquist AB, Silk BJ, et al. Multistate outbreak of listeriosis associated with cantaloupe. N Engl J Med 2013;369:944-53. [Crossref] [PubMed]
- Maury MM, Tsai YH, Charlier C, et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet 2016;48:308-13. [Crossref] [PubMed]
- Wang H, Luo L, Zhang Z, et al. Prevalence and molecular characteristics of Listeria monocytogenes in cooked products and its comparison with isolates from listeriosis cases. Front Med 2018;12:104-12. [Crossref] [PubMed]
- Jamali H, Paydar M, Ismail S, et al. Prevalence, antimicrobial susceptibility and virulotyping of Listeria species and Listeria monocytogenes isolated from open-air fish markets. BMC Microbiol 2015;15:144. [Crossref] [PubMed]
- Lotfollahi L, Chaharbalesh A, Ahangarzadeh Rezaee M, et al. Prevalence, antimicrobial susceptibility and multiplex PCR-serotyping of Listeria monocytogenes isolated from humans, foods and livestock in Iran. Microb Pathog 2017;107:425-9. [Crossref] [PubMed]


