
Of fungi and men: role of fungi in pancreatic cancer carcinogenesis
Pancreatic ductal adenocarcinoma (PDAC) is a highly malignant tumor with a 5-year survival rate of 9% (1). This poor survival rate is caused by the late discovery of the disease, the early metastatic spread and the resistance to most currently available treatments (2). Early detection and accurate diagnosis are critical to improving outcome, as clearly demonstrated by a recent study which reported a 5-year overall survival above 80% for patients in stage IA (3). Although several detection methods, such as analyses of plasma proteins, are widely used in the diagnosis of tumors, they have very limited impact on the early detection of PDAC (4).
Research on key factors in PDAC carcinogenesis that might be used as biomarkers for early detection is therefore urgently needed. Many factors have so far been associated with increased risk of PDAC, namely, smoking, heavy alcohol use, diabetes mellitus, obesity, family history and inherited genetic variation (5). Recent studies suggest the involvement of components of tumor microenvironment (TME) which select PDAC for different fitness advantages when encountering extracellular matrix components, immune cells and restricted nutrient or oxygen gradients (6).
The TME of PDAC contains diverse bacterial species (7,8), which have been associated with carcinogenesis (9), as well as to the patient’s outcome (10). All these studies did not investigate the potential role of other germs, including fungi.
However, a recent publication revealed for the first time that the fungal mycobiome promotes pancreatic oncogenesis (11). In this seminal study, the abundance of intrapancreatic fungi and fungal DNA content in PDAC was compared to normal pancreatic tissues, using Fluorescent in situ hybridization and quantitative PCR techniques. These analyses showed a marked increase in intratumoral fungi in PDAC samples obtained from mouse models and human. Next, Aykut and colleagues assessed the mycobiome profiles of PDAC using 18S internal transcribed space sequencing, and discovered that the mycobiome composition of cancerous tissues was distinct from that of the gut or the normal pancreas, both in human and mice specimens. Of note, Malassezia was the most prevalent fungal genus in PDAC. These results suggest fungal dysbiosis during tumorigenesis, which was further investigated (Figure 1). Ablation of mycobiome with antifungal medications, such as amphotericin B or fluconazole, protected mice against oncogenic progression. When mice treated with amphotericin B were repopulated with Malassezia an accelerated growth of pancreatic tumors was observed. Conversely, repopulation with Candida spp., Saccharomyces cerevisiae or Aspergillus spp. did not affect the growth of the tumors.
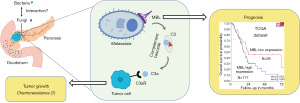
Interestingly, the same research group has previously demonstrated involvement of the microbiome in the development of PDAC using the same mice models and similar methodologies (9). They found that PDAC harbors a considerably more abundant microbiome than normal pancreas in both mice and humans. Proteobacteria and Actinobacteria were more abundant in PDAC than in gut and were associated with PDAC oncogenesis. Ablation of the microbiome protected the mice against preinvasive and invasive neoplastic lesions, whereas introduction of the bacteria from PDAC-bearing hosts reversed this protection.
The association of bacteria with PDAC oncogenesis has been confirmed by several other studies (8,10), but the dominant bacterial species in PDAC varied among these studies. Moreover, some criticisms were raised about how critical clinically-relevant factors could influence the microbiome composition, including usage of proton pump inhibitors and the occurrence of biliary obstruction, which is often palliated preoperatively by endoscopic preoperative biliary drainage (12).
Since the human microbiota generally consists of bacteria, archaea, viruses and fungi which form a highly complex network, it is likely that the fungi and bacteria in TME promote the progression of PDAC through similar molecular mechanisms or even synergistically. Thus, it would be interesting to study how the interactions between these fungi and bacteria influence tumorigenesis.
Notably, Aykut and colleagues investigated how Malassezia promotes PDAC growth. Malassezia is known to be able to bind C-type lectin receptors (13,14). A thoughtful analysis of the transcriptomic data from The Cancer Genome Atlas unraveled the correlation between high levels of expression of the mannose-binding lectin, MBL, and significantly shorter survival of PDAC patients (Figure 1). MBL is a soluble recognition molecule that binds to terminal sugar residues present on the surface of microorganisms. Upon MBL binding, complement elements C2 and/or C4 can be activated through serine-related proteases (15). Keeping with these findings, mice lacking MBL exhibited delayed oncogenic progression. Moreover, the presence of Malassezia could not accelerate the PDAC progression in the MBL-null mice. The central complement component C3, downstream of MBL, was also considered a key determinant in PDAC oncogenesis. Expression of C3 was indeed associated with shorter survival of PDAC patients and the mice lacking C3 displayed reduced tumor growth. Collectively, these results indicate that fungi were able to promote PDAC progression via MBL-C3 pathway activation (Figure 1). However, further studies should investigate whether other species of fungi, such as Candida spp., S. cerevisiae and Aspergillus spp. could not induce PDAC progression due to a differentiation in immune recognition.
Remarkably, bacteria seem to promote progression of tumor growth via different mechanisms. The previous study by Pushalkar and collaborators (9) showed that dysbiosis of the bacterial microbiome accelerated growth of PDAC by suppressing both innate and adaptive immunity via TLR2 and TLR5 recognition. Some bacterial species in the microbiome, however, may induce beneficial effects. Riquelme and collaborators indeed (10) discovered 3 tumor bacterial taxa—Saccharopolyspora, Pseudoxanthomonas, and Streptomyces—highly enriched in PDAC patients who experienced very long survival. These bacterial species contribute to the recruitment and activation of CD8 T cells to the TME and hence induce a more effective antitumor response. Similar “positive effects” might be detected also with some fungi species.
However, when the presence of both fungi and bacteria in TME is considered, the mechanisms for the tumorigenesis may be more complicated. On one hand, the C3 activated by the binding of fungi to MBL could modulate the bacteria-suppressed innate and adaptive immunities and thereby the function of T cells. On the other hand, immunosuppression caused by bacteria may complicate the fungi-related MBL-C3 axis (16). Therefore, future studies should investigate the potential mechanisms underlying different effects caused by the interplay between bacteria and fungi.
Aykut and collaborators (11) also examined the source of fungi. Using a green-fluorescent-protein (GFP) labeled strain, the authors established that fungi are able to translocate into pancreas from gut within 30 minutes. Similarly, a GFP-labelled Enterococcus faecalis strain was found to migrate into the pancreas in 30 minutes (9). As we know, the human gastrointestinal tract hosts a complex microbiota. Although the number and abundance of fungi in this gut community are orders of magnitude lower than those of bacteria (17), the function of fungi was equivalent to that of bacteria. Similar to the dysbiosis of microbiome, dysbiosis of the mycobiome has been associated with various gut-related diseases (18) and systemic diseases, such as neurological diseases (19). Fungal genera commonly associated with the human gut are Candida, Saccharomyces, Malassezia, Aspergillus and Pichia. Although research on the mycobiome in health and disease is still in its infancy, existing data suggest that reduced mycobiome species richness and enrichment of certain fungal species, such as Candida albicans, Saccharomyces cerevisiae and Malassezia can lead to the onset of systemic diseases (19).
Another human microbiota worth mentioning is the oral microbiota. It has been suggested that part of the intestinal bacteria and fungi are of oral origin, with more than 45% overlap in the microbiota between oral cavity and gut (20,21). The increase of PDAC risk has been associated with a very common oral infection: periodontal disease (22). The levels of two periodontal pathogens, Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans, were elevated in PDAC patients (23). Oral bacteria are thought to reach the pancreas by swallowing or via the circulatory system after mastication and personal oral hygiene (24). Being an important component of oral microbiota, oral fungi may also play a role in the progression of PDAC. Interestingly, the PDAC related Malassezia genus is an overlooked oral commensal as it was recently found in high prevalence and abundance in saliva of healthy individuals (25), although it was previously considered as commensal and pathogen of the skin and lungs.
Aykut and collaborators also reported that the ablation of mycobiome affected the activity of gemcitabine (11). Gemcitabine is still ranking among the most widely prescribed drugs in PDAC patients, but the response to this treatment is extremely poor (2). Strikingly, Geller and collaborators described several bacterial species, such as Gammaproteobacteria, which express the enzyme cytidine deaminase, whose long-form (CDDL) was shown to metabolize gemcitabine into its inactive metabolite. Interestingly, preclinical models injected with bacterial CDDL displayed a reduced response to gemcitabine, but this resistance was neutralized by the antibiotic ciprofloxacin. The increased knowledge on the mycobiome in pancreatic tissues, as well as its role in chemoresistance, could then provide also innovative therapeutic strategies.
In conclusion, the study of Aykut and collaborators reveals that dysbiosis of mycobiome in TME can promote the progression of PDAC via MBL-C3 axis, supporting further studies on these candidates as new biomarkers for early diagnostics. Moreover, when taking into account the previous data on the involvement of the microbiome in the progression and drug sensitivity of PDAC, measures that are able to alter the composition of both microbiome and mycobiome might provide novel therapeutic approaches to slow down cancer progression and overcome chemoresistance.
Acknowledgments
Funding: This work was supported by grants from the Cancer Center Amsterdam (CCA) Foundation, Associazione Italiana per la Ricerca sul Cancro (AIRC) and Fondazione Pisana per la Scienza (FPS).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Le Li (Department of Pancreatic and Biliary Surgery, The First Affiliated Hospital of Harbin Medical University, Harbin Medical University, Harbin, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2723). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- American Cancer Society. Cancer Facts & Figures 2020. Atlanta Ga: American Cancer Society, 2020.
- Giovannetti E, van der Borden CL, Frampton AE, et al. Never Let It Go: Stopping Key Mechanisms Underlying Metastasis to Fight Pancreatic Cancer. Semin Cancer Biol 2017;44:43-59. [Crossref] [PubMed]
- Blackford AL, Canto MI, Klein AP, et al. Recent Trends in the Incidence and Survival of Stage 1A Pancreatic Cancer: A Surveillance, Epidemiology, and End Results Analysis. J Natl Cancer Inst 2020. [Epub ahead of print]. [Crossref] [PubMed]
- Root A, Allen P, Tempst P, et al. Protein Biomarkers for Early Detection of Pancreatic Ductal Adenocarcinoma: Progress and Challenges. Cancers 2018;10:67. [Crossref] [PubMed]
- Pereira SP, Oldfield L, Ney A, et al. Early Detection of Pancreatic Cancer. Lancet Gastroenterol Hepatol 2020;5:698-710. [PubMed]
- Makohon-Moore A, Iacobuzio-Donahue CA. Pancreatic Cancer Biology and Genetics from an Evolutionary Perspective. Nat Rev Cancer 2016;16:553-65. [Crossref] [PubMed]
- Mitsuhashi K, Nosho K, Sukawa Y, et al. Association of Fusobacterium Species in Pancreatic Cancer Tissues with Molecular Features and Prognosis. Oncotarget 2015;6:7209-20. [Crossref] [PubMed]
- Geller LT, Baezily-Rokni M, Danino T, et al. Potential Role of Intratumor Bacteria in Mediating Tumor Resistance to the Chemotherapeutic Drug Gemcitabine. Science 2017;357:1156-60. [Crossref] [PubMed]
- Pushalkar S, Hundeyin M, Daley D, et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 2018;8:403-16. [Crossref] [PubMed]
- Riquelme E, Zhang Y, Zhang L, et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019;178:795-806.e12. [Crossref] [PubMed]
- Aykut B, Pushalkar S, Chen R, et al. The Fungal Mycobiome Promotes Pancreatic Oncogenesis via Activation of MBL. Nature 2019;574:264-7. [Crossref] [PubMed]
- Meijer LL, Zwart ES, Brandt BW, et al. Tumor Microbiome: Pancreatic Cancer and Duodenal Fluids Contain Multitudes, …but Do They Contradict Themselves? Crit Rev Oncol Hematol 2019;144:102824. [Crossref] [PubMed]
- Yamasaki S, Matsumoto M, Takeuchi O, et al. C-Type Lectin Mincle is an Activating Receptor for Pathogenic Fungus, Malassezia. Proc Natl Acad Sci U S A 2009;106:1897-902. [Crossref] [PubMed]
- Ishikawa T, Itoh F, Yoshida S, et al. Identification of Distinct Ligands for the C-Type Lectin Receptors Mincle and Dectin-2 in the Pathogenic Fungus Malassezia. Cell Host Microbe 2013;13:477-88. [Crossref] [PubMed]
- Thiel S, Vorup-Jensen T, Stover CM, et al. A Second Serin Protease Associated with Mannan-Binding Lectin that Activates Complement. Nature 1997;386:506-10. [Crossref] [PubMed]
- Kolev M, Markiewski MM. Targeting Complement-Mediated Immunoregulation for Cancer Immunotherapy. Semin Immunol 2018;37:85-97. [Crossref] [PubMed]
- Rowan-Nash AD, Korry BJ, Mylonakis E, et al. Cross-Domain and Viral Interactions in the Microbiome. Microbiol Mol Biol Rev 2019. [Crossref] [PubMed]
- Sam QH, Chang MW, Chai LYA. The Fungal Mycobiome and Its Interaction with Gut Bacteria in the Host. Int J Mol Sci 2017;18:330. [Crossref] [PubMed]
- Enaud R, Vandenborght LE, Coron N, et al. The Mycobiome: A Neglected Component in the Microbiota-Gut-Brain Axis. Microorganisms 2018;6:22. [Crossref] [PubMed]
- Segata N, Haake SK, Mannon P, et al. Composition of the Adult Digestive Tract Bacterial Microbiome Based on Seven Mouth Surfaces, Tonsils, Throat and Stool Samples. Genome Biol 2012;13:R42. [Crossref] [PubMed]
- Klimesova K, Jiraskova Zakostelska Z, Tlaskalova-Hogenova H. Oral Bacterial and Fungal Microbiome Impacts Colorectal Carcinogenesis. Front Microbiol 2018;9:774. [Crossref] [PubMed]
- Michaud DS, Joshipura K, Giovannucci E, et al. A Prospective Study of Periodontal Disease and Pancreatic Cancer in US Male Health Professionals. J Natl Cancer Inst 2007;99:171-5. [Crossref] [PubMed]
- Michaud DS, Izard J, Wilhelm-Benartzi CS, et al. Plasma Antibodies to Oral Bacteria and Risk of Pancreatic Cancer in a Large European Prospective Cohort Study. Gut 2013;62:1764-70. [Crossref] [PubMed]
- Choy ATF, Carnevale I, Coppola S, et al. The Microbiome of Pancreatic Cancer: from Molecular Diagnostics to New Therapeutic Approaches to Overcome Chemoresistance Caused by Metabolic Inactivation of Gemcitabine. Expert Rev Mol Diagn 2018;18:1005-9. [Crossref] [PubMed]
- Diaz PI, Hong BY, Dupuy AK, et al. Mining the Oral Mycobiome: Methods Components, and Meaning. Virulence 2017;8:313-23. [Crossref] [PubMed]


