Methylation-driven genes and their prognostic value in cervical squamous cell carcinoma
Introduction
Cervical cancer is prevalent in women, causing 530,000 new cases and 270,000 deaths worldwide annually (1). Cervical squamous cell carcinoma (CESC) is the most common histological subtype of cervical cancer. The prognosis of CESC remains poor due to its high metastasis and recurrence. Cervical cancer has a wide spectrum of high-risk factors are multiple, like human papilloma virus infection and unprotected sex (2). New molecular technology has been used to dig out an increasing number of CESC-related oncogenes, such as PTE (3), MYC (4) and TP53 (5). However, no effective therapeutic drugs have been developed based on these target genes.
DNA methylation has been proved to regulate carcinogenesis-associated cellular processes (6), which provides a possibility to find therapeutic targets through assessing their DNA methylation. In recent years, increasing evidence has shown that the occurrence and progression of CESC are modulated by diverse mechanisms, including the methylation of CESC-associated genes (7).
Bioinformatic analysis is widely used to explore the molecular mechanism and candidate biomarkers in cancers. The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/publications/, publication guidelines) provides genetic and epigenetic profiles. The MethylMix algorithm is used to screen out disease-specific methylation-driven genes based on a β-mixed model (8,9). In the present study, we screened out the methylation-driven genes and assessed their prognostic value in CESC.
Methods
Data collection and analysis
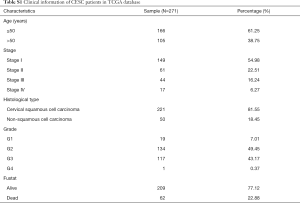
We downloaded the expression profiles of mRNAs (level 3) in 309 cases (306 tumor tissues and 3 normal tissues), the methylation data (level 3) in 312 cases (309 tumor tissues and 3 normal tissues) from TCGA database (http://cancergenome.nih.gov/). The sequenced data were obtained from Illumina HiSeqRNASeq and Illumina Human Methylation 450 platform. The corresponding clinical information of EC patients was also downloaded from TCGA database and showed in Table S1. First of all, these data were normalized and differentially expressed genes (DEGs) and aberrant methylated genes were screened out using LIMMA package (10). Next, the correlation between gene methylation and gene expression was computed via MethylMix algorithm. We identified DEGs in CESC and the CESC-specific methylation-driven genes through the β-mixed model.
Full table
Functional annotations of methylation-driven genes
The biological functions of these methylation-driven genes were further investigated based on The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (http://david.abcc.ncifcrf.gov/) (11). The Gene Ontology (GO) enrichment analysis results were plotted in the GOplot R package (https://cran.r-project.org/web/packages/GOplot/) (12). In addition, we used ConsensusPathDB (http://cpdb.molgen.mpg.de/) online software to perform Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (13). P<0.05 was set as the cutoff criterion in GO and KEGG analyses.
Construction of protein–protein interaction (PPI)
To evaluate the interrelationships among these methylation-driven genes, we uploaded them onto the Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org/) to construct a PPI network (14).
Construction of risk model
To further screen out prognosis-related methylation-driven genes of CESC, we performed univariable and multivariate Cox analyses, and then constructed a linear risk model (15). The prognostic score = expRNA1×βRNA1+expRNA2×βRNA2+expRNA3×βRNA3+..expRNAn×βRNAn (expRNA is the expression level of each methylation-driven gene, and βRNA is the regression coefficient calculated by the multivariate Cox regression analysis). The patients were divided into two groups (high-risk and low-risk) according to the median risk value. The Kaplan-Meier survival analysis was conducted to compare the overall survival rate of the two groups in survival R package (the log-rank P<0.05). A 5-year-dependent receiver operating characteristic (ROC) curve was further plotted to evaluate the predictive value of the risk model. In addition, univariate and multivariate Cox regression analyses were used to determine whether the candidate driver genes were independent risk factors. Meanwhile, the stratification analysis was operated based on clinicopathological features such as patient’s age, tumor grade, stage and histological type.
Survival analysis of candidate driver genes and mapping of methylation sites
Using Survival R package, a survival analysis was performed to explore the correlation between the methylation and expression of key prognosis-associated genes in CESC. We extracted relevant methylated sites of key genes from the CESC methylation data. |Cor| >0.35 was used as cutoff criterion (16).
Gene set enrichment analysis (GSEA) and pathways enriched analysis
In TCGA set validation, 306 CESC samples were divided into highly-expressed and lowly-expressed groups according to the median expression level of identified genes. The biological functions of the key methylation-driven genes were clarified with GSEA (http://software.broadinstitute.org/gsea/index.jsp) (17). Annotated gene sets of c2.cp.kegg.v6.0.symbols.gmt in Molecular Signatures Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb/index.jsp) were chosen as the references in GSEA software (18). The Nom. P<0.0001 and the enrichment score (ES) ≥0.70 were chosen as the cutoff criterion.
Construction and validation of the nomogram
A nomogram and a calibrate curve were plotted by the “rms” package on R. The accuracy of the nomogram was examined using the consistency between the actual and the predicted outcomes. Then, we submitted these outcomes to the calibration curve to visualize the performance of the nomogram. The 45° line represented the best prediction (19).
Results
Aberrant methylation-driven genes in CESC
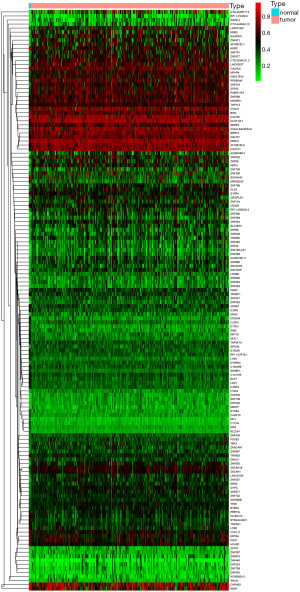
The gene expression data of 309 samples (306 CESC samples and 3 normal samples) and the methylation data of 312 samples (309 CESC samples and 3 normal samples) were downloaded from TCGA. The genes with abnormal expression were found out based on the LIMMA software package. The correlation between methylation level and expression level of each gene was investigated based on the MethylMix package. In the Wilcoxon rank test, the differential methylation was defined with |log Fold change (FC)| >0 and adjusted P<0.05, |Cor| >0.45. As a result, 144 CESC-related methylation-driven genes were screened out, and the heat maps of these aberrant methylation-driven genes were shown in Figure 1.
Annotations and pathways associated with methylation-driven genes in CESC
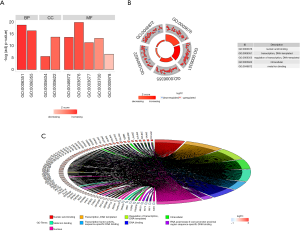
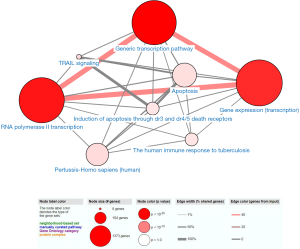
We performed GO and KEGG analyses by using DAVID and ConsensusPathDB online software, respectively. GO analysis revealed that in the aspect of BP, the genes were significantly enriched in “Transcription, DNA-templated and regulation of transcription, DNA-templated”. In the aspect of MF group, the genes were mainly involved in “Nucleic acid binding, Metal ion binding, Transcription factor activity, sequence-specific DNA binding, DNA binding and RNA polymerase II core promoter proximal region sequence-specific DNA binding”. In the aspect of CC, the genes were mainly enriched in “Nucleus” (Figure 2). KEGG pathway analysis showed that the genes were mainly enriched in “Generic transcription pathway, RNA polymerase II transcription, Gene expression (Transcription), The human immune response to tuberculosis, Apoptosis, Induction of apoptosis through dr3 and dr4/5 death receptors and TRAIL signaling” (Figure 3).
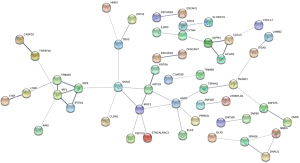
Construction of PPI network
The PPI network contained 124 nodes and 74 edges (Figure S1).
Prognostic risk model based on CESC methylation-driven genes
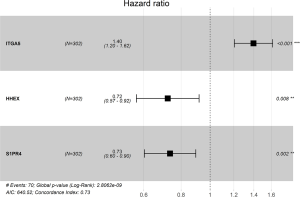
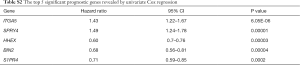
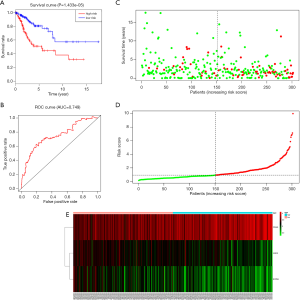
Through univariable and multivariate Cox regression analyses, we screened out prognosis-associated methylation-driven genes (Table S2). Then, we further performed a multivariate Cox proportional hazards regression analysis based on the top five most associative genes (ITGA5, SPRY4, HHEX, BIN2 and S1PR4). The results showed that the risk score calculated by three genes (ITGA5, HHEX and S1PR4) could be used as an independent indicator of CESC prognosis. The risk score = (0.334 × ITGA5) + (−0.326 × HHEX) + (−0.308 × S1PR1) (Figure S2). Moreover, according to their median values, a total of 302 CESC samples with corresponding clinical samples were divided into a high-risk group (151 samples) and a low-risk group (151 samples). And the results of Kaplan-Meier survival analysis showed that the overall survival rate in the low-risk group was significantly higher than that in the high-risk group (Figure 4A). The 5-year overall survival ROC curve showed the area under curve (AUC) was 0.749 (Figure 4B). It suggested that this model was reliable to evaluate 5-year overall survival of patients with CESC. Meanwhile, the risk scores, survival time, and expression levels indicated by the three genes for each patient were showed in Figure 4C,D,E. The expression levels of the three genes in low-risk and high-risk groups were shown in Figure S3A. The heatmap showed the expression of the three genes in high- and low-risk patients. We observed significant differences in histological type (P<0.01) and tumor status (P<0.001) between the high- and low-risk groups (Figure S3B).
Full table

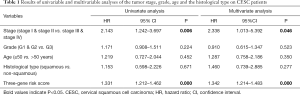
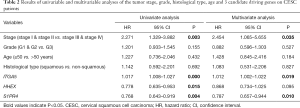
Moreover, we further performed univariate and multivariate Cox regression analyses of the 3-generiskscore, stage (stage I & stage II vs. stage III & stage IV), grade (G1 & G2 vs. G3), and age (≤50 vs. >50). As shown in Tables 1,2, tumor stage (HR =2.338, 95% CI: 1.013–5.392, P value =0.046) and the risk score (HR =1.342, 95% CI: 1.214–1.483, P value =0.000) were found as independent prognostic factors for CESC patients. ITGA5 (HR =1.012, 95% CI: 1.002–1.022, P value =0.019) and S1PR4 (HR =0.787, 95% CI: 0.657–0.944, P value =0.010), stage (HR =2.454, 95% CI: 1.065–5.655, P value =0.035) were also identified as independent prognostic factors by further univariable and multivariable analyses of the tumor stage, grade, and expression of three candidate driving gene.
Full table
Full table
Survival analysis and mapping of methylation sites
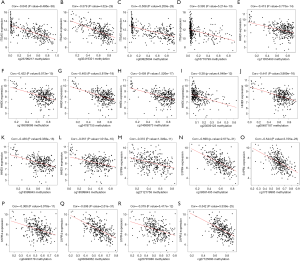
Through a joint Kaplan-Meier survival analysis, we found that the patients with lowly-expressed and hyper-methylated ITGA5 presented a better prognosis than those with highly-expressed and hypo-methylated ITGA5. Conversely, those with highly-expressed and hypomethylated HHEX and S1PR4 presented higher overall survival rate than those with lowly-expressed and hypermethylated HHEX and S1PR4 (Figure 5). In addition, the association between the methylation and the expression of the three genes was shown in Figure 6.

Enriched biological processes and pathways of key methylation-driven genes
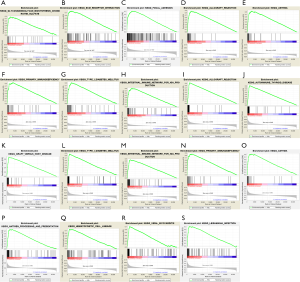
GSEA was conducted to explore the biological functions enriched in each gene. ITGA5 was mostly enriched in “Glycosaminoglycan biosynthesis chondroitin sulfate, ECM receptor interaction and Focal adhesion” (Figure S4A,B,C); HHEX in “Allograft rejection, Asthma, Primary immunodeficiency, Type I diabetes mellitus and Intestinal immune network for IgA production” (Figure S4D,E,F,H); S1PR4 in “Allograft rejection, Autoimmune thyroid disease, Graft versus host disease, Type I diabetes mellitus, Intestinal immune network for IgA production, Primary immunodeficiency, Asthma, antigen processing and presentation, Hematopoietic cell lineage, Viral myocarditis and Leishmania infection” (P<0.0001) (Figure S4I-S).
Accuracy of nomogram
We created a nomogram to estimate the 1-, 3- and 5-year OS. The five independent prognostic factors included stage, age, histological type, grade and risk score (Figure 7A). The 45° line represented the best prediction. Calibration plots suggested that the nomogram performed well to predict the 1-year OS (Figure 7B,C,D). ROC curve analysis showed that the AUC value of the risk model was 0.730, much significantly higher than that of clinical stage (AUC =0.651), grade (AUC =0.621), age (AUC =0.573) and histological type (AUC =0.512). The nomogram was also efficient in predicting 3- and 5-year OS (Figure 7E,F,G).
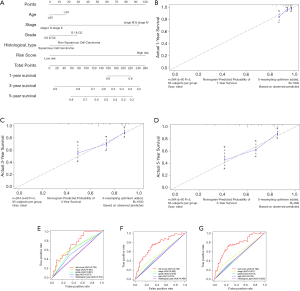
Prognostic value and clinical utility of hub genes
The stratification analysis was performed based on grade, age, stage and histological type. The patients were stratified into grade I/II subgroups, stage I/II and stage III/IV subgroups. As shown in Figure 8A, the prognosis of high-risk patients was significantly worse than that of low-risk patients in the grade I/II, stage I/II and stage III/IV subgroups (Figure 8B,C). We also assessed the prognostic ability of risk signature combined with age and histological type. The patients were also stratified into >50 years subgroup and ≤50 years subgroups. Interestingly, we found that high-risk patients in two subgroups were inclined to unfavorable OS (Figure 8D,E). The similar trend was also observed in the squamous cancer subgroup (Figure 8F).
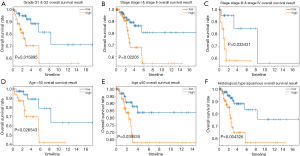
Discussion
CESC can be caused by abnormal genetic and epigenetic changes (20). Recently, increasing methylation-driven genes have been identified as candidate biomarkers in cancers, such as esophageal squamous cell carcinoma, lung squamous cell carcinoma and renal clear cell carcinoma (21-23). In the present study, we screened out methylation-driven genes associated with CESC.
Firstly, 144 methylation-driven genes were identified. GO analysis revealed that these genes were mostly associated with “Nucleic acid binding, Transcription, DNA-templated and Regulation of transcription, DNA-templated”. KEGG analysis showed that they were mainly involved in the pathways including “Generic transcription pathway, RNA polymerase II transcription, Gene expression (Transcription)”. The gene expression programs are governed by thousands of transcriptional factors and chromatin regulators. The mutations and epigenetic modification of these regulators contribute to tumorigenesis. Our findings indicated that most of these methylation-driven genes might regulate the transcription and expression of oncogenes/anti-oncogene in CESC. In addition, through performing univariable and multivariate Cox analyses, three candidate driver genes (ITGA5, HHEX and S1PR4) were identified as CESC prognostic-related genes and used to construct a risk model. We divided the CESC samples into two groups. Significant difference was showed in the overall survival between the two groups, suggesting that the risk model was effective in predicting the prognosis of CESC patients. The AUC of 5-year overall survival ROC curve was 0.749, suggesting that the risk model is accurate in predicting the prognosis of CESC patients. These findings were also validated by our nomogram. We further found that the three-genes-based risk score and the TNM stage might be independent prognostic factors. However, further studies are needed to support our findings.
ITGA5, as a member of integrin family, has been reported to mediate the initial adhesion process in cancers, such as the ovarian cancer (24), and colorectal cancer (25). In recent years, the role of integrins in cancers has been illustrated. Various integrin inhibitors have proved their efficacy in resisting cancer progression (26). However, we found that hyper-methylated and lowly-expressed ITGA5 was associated with a better prognosis of CESC, which is consistent with the finding of Fang et al., in that ITGA5 expression was down-regulated in invasive breast cancer cells by increasing methylation in the promotor of ITGA5 (27). HHEX is down-regulated in multiple malignancies (28). Down-regulation of HHEX suppresses the development of breast cancer, thyroid cancer and hepatocellular carcinoma (29-31). In our present study, we also found hypermethylation and low-expression of HHEX were associated with a poor prognosis in CESC.S1PRs family was found highly-expressed in multiple cancers, and associated with poor prognosis of cancers in a previous study (32). Inversely, we found that the hypermethylation and low-expression of gene S1PR4 was associated with a poor prognosis of CESC, suggesting that S1PR4 might restrain the progression of CESC. However, up to date, all the performances of three genes have not been experimentally validated. GSEA further revealed the biological functions of ITGA5, HHEX and S1PR4. Thus, we speculated that ITGA5 might participate in the progression of CESC by mediating pathways, including glycosaminoglycan biosynthesis chondroitin sulfate, ECM receptor interaction and Focal adhesion. HHEX and S1PR4 were mostly associated with autoimmune processes. The relationship between tumorigenesis and autoimmunity has been confirmed in recent years, especially in paraneoplastic syndromes (33), indicating that immunotherapy might be effective in treating cancers.
In conclusion, our study defined three methylation-driven genes (ITGA5, HHEX and S1PR4) associated with CESC development and built a 3-gene risk signature for predicting cancer prognosis. However, biofunctions of these genes remain to be unveiled with more in-depth research. The three genes might serve as potential prognostic biomarkers and therapeutic targets in the treatment of CESC.
Acknowledgments
Funding: National Natural Science Foundation (81472442, 81872119) and Jiangsu province medical innovation team (CXTDA2017008) supported our study.
Footnote
Peer Review File: Available at http://dx.doi.org/10.21037/atm-19-4577
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-19-4577). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The statement of ethics approval and the informed consent were not necessary in this study, because our present study was a bioinformatics analysis based on TCGA database, and all the sample information was downloaded from open databases.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Small W Jr, Bacon MA, Bajaj A, et al. Cervical cancer: A global health crisis. Cancer 2017;123:2404-12. [Crossref] [PubMed]
- Chelimo C, Wouldes TA, Cameron LD, et al. Risk factors for and prevention of human papillomaviruses (HPV), genital warts and cervical cancer. J Infect 2013;66:207-17. [Crossref] [PubMed]
- Vázquez-Ulloa E, Lizano M, Aviles-Salas A, et al. Abnormal distribution of hDlg and PTEN in premalignant lesions and invasive cervical cancer. Gynecol Oncol 2011;122:663-8. [Crossref] [PubMed]
- Yuan H, Krawczyk E, Blancato J, et al. HPV positive neuroendocrine cervical cancer cells are dependent on Myc but not E6/E7 viral oncogenes. Sci Rep 2017;7:45617. [Crossref] [PubMed]
- Klug SJ, Wilmotte R, Santos C, et al. TP53 polymorphism, HPV infection, and risk of cervical cancer. Cancer Epidemiol Biomarkers Prev 2001;10:1009-12. [PubMed]
- Klutstein M, Nejman D, Greenfield R, et al. DNA Methylation in Cancer and Aging. Cancer Res 2016;76:3446-50. [Crossref] [PubMed]
- Bhat S, Kabekkodu SP, Noronha A, et al. Biological implications and therapeutic significance of DNA methylation regulated genes in cervical cancer. Biochimie 2016;121:298-311. [Crossref] [PubMed]
- Gevaert O. MethylMix: an R package for identifying DNA methylation-driven genes. Bioinformatics 2015;31:1839-41. [Crossref] [PubMed]
- Gevaert O, Tibshirani R, Plevritis SK. Pancancer analysis of DNA methylation-driven genes using MethylMix. Genome Biol 2015;16:17. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 2015;31:2912-4. [Crossref] [PubMed]
- Kamburov A, Stelzl U, Lehrach H, et al. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res 2013;41:D793-800. [Crossref] [PubMed]
- Szklarczyk D, Jensen LJ. Protein-protein interaction databases. Methods Mol Biol 2015;1278:39-56. [Crossref] [PubMed]
- Gao C, Zhuang J, Li H, et al. Exploration of methylation-driven genes for monitoring and prognosis of patients with lung adenocarcinoma. Cancer Cell Int 2018;18:194. [Crossref] [PubMed]
- Gao C, Zhuang J, Zhou C, et al. Prognostic value of aberrantly expressed methylation gene profiles in lung squamous cell carcinoma: A study based on The Cancer Genome Atlas. J Cell Physiol 2019;234:6519-28. [Crossref] [PubMed]
- Croken MM, Qiu W, White MW, et al. Gene Set Enrichment Analysis (GSEA) of Toxoplasma gondii expression datasets links cell cycle progression and the bradyzoite developmental program. BMC Genomics 2014;15:515. [Crossref] [PubMed]
- Liberzon A, Birger C, Thorvaldsdottir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- Pencina MJ, D'Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med 2004;23:2109-23. [Crossref] [PubMed]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726-34. [Crossref] [PubMed]
- Lu T, Chen D, Wang Y, et al. Identification of DNA methylation-driven genes in esophageal squamous cell carcinoma: a study based on The Cancer Genome Atlas. Cancer Cell Int 2019;19:52. [Crossref] [PubMed]
- Li Y, Gu J, Xu F, et al. Novel methylation-driven genes identified as prognostic indicators for lung squamous cell carcinoma. Am J Transl Res 2019;11:1997-2012. [PubMed]
- Wang J, Zhao H, Dong H, et al. LAT, HOXD3 and NFE2L3 identified as novel DNA methylation-driven genes and prognostic markers in human clear cell renal cell carcinoma by integrative bioinformatics approaches. J Cancer 2019;10:6726-37. [Crossref] [PubMed]
- Ohyagi-Hara C, Sawada K, Kamiura S, et al. miR-92a inhibits peritoneal dissemination of ovarian cancer cells by inhibiting integrin alpha5 expression. Am J Pathol 2013;182:1876-89. [Crossref] [PubMed]
- Yoo HI, Kim BK, Yoon SK. MicroRNA-330-5p negatively regulates ITGA5 expression in human colorectal cancer. Oncol Rep 2016;36:3023-9. [Crossref] [PubMed]
- Raab-Westphal S, Marshall JF, Goodman SL. Integrins as Therapeutic Targets: Successes and Cancers. Cancers (Basel) 2017;9:110. [Crossref] [PubMed]
- Fang Z, Yao W, Xiong Y, et al. Functional elucidation and methylation-mediated downregulation of ITGA5 gene in breast cancer cell line MDA-MB-468. J Cell Biochem 2010;110:1130-41. [Crossref] [PubMed]
- Gaston K, Tsitsilianos MA, Wadey K, et al. Misregulation of the proline rich homeodomain (PRH/HHEX) protein in cancer cells and its consequences for tumour growth and invasion. Cell Biosci 2016;6:12. [Crossref] [PubMed]
- Puppin C, Puglisi F, Pellizzari L, et al. HEX expression and localization in normal mammary gland and breast carcinoma. BMC Cancer 2006;6:192. [Crossref] [PubMed]
- D'Elia AV, Tell G, Russo D, et al. Expression and localization of the homeodomain-containing protein HEX in human thyroid tumors. J Clin Endocrinol Metab 2002;87:1376-83. [Crossref] [PubMed]
- Su J, You P, Zhao JP, et al. A potential role for the homeoprotein Hhex in hepatocellular carcinoma progression. Med Oncol 2012;29:1059-67. [Crossref] [PubMed]
- Wang C, Mao J, Redfield S, et al. Systemic distribution, subcellular localization and differential expression of sphingosine-1-phosphate receptors in benign and malignant human tissues. Exp Mol Pathol 2014;97:259-65. [Crossref] [PubMed]
- Mitchell WG, Blaes F. Cancer and Autoimmunity: Paraneoplastic Neurological Disorders Associated With Neuroblastic Tumors. Semin Pediatr Neurol 2017;24:180-8. [Crossref] [PubMed]