
Identification of critical genes in gastric cancer to predict prognosis using bioinformatics analysis methods
Introduction
As one of the most heterogeneous, multifactorial diseases in the world, gastric cancer (GC) persists in its high morbidity and mortality, especially in some areas of China (1). It is damaging to human physical and mental health, and thus also aggravates the public health and economic burden of China (2,3). Even though the swift development of technologies like gastroscopy and computed tomography has enabled early diagnosis and treatment to greatly slow the deleterious progression of GC, some patients still suffer from unusual patterns of locoregional and systemic recurrence. Furthermore, a large portion of patients still progress to the advanced stage of GC for various reasons. For these reasons, the overall survival (OS) of GC patients remains low around the world. Indeed, GC, as the fourth most common global malignancy, also ranks as the world’s second leading cause of cancer-related death (4,5). Alarmingly, the incidence of GC is gradually increasing in young people (6). Even though some prognostic biomarkers have been identified and applied in clinical treatment (7), there is a still an urgent need to search for other significant genes in GC in order to better understand the underlying mechanism and improve the treatment effect. Gene chip has been used for several decades and has proven to be a reliable technique. Using it, differentially expressed genes (DEGs) can be detected quickly, and gene chip may even be used to produce many genomic information for storage in public databases. This wealth of data could then be exploited as a base for a large number of investigative studies. Indeed, an increasing amount of bioinformatical studies on GC have been conducted, which gives assurance that the underlying mechanisms of GC can be explored with integrated bioinformatical methods.
In this study, we first selected GSE33335 and GSE63089 from Gene Expression Omnibus (GEO). Secondly, the DEGs in the two datasets above were identified by using the GEO2R online tool and Venn diagram software. Thirdly, we further analyzed those DEGs containing Gene ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway by using the Database for Annotation, Visualization and Integrated Discovery (DAVID). Fourthly, a protein-protein interaction (PPI) network and Cytotype Molecular Complex Detection (MCODE) were established for further analysis of the DEGs which enabled us to obtain several core genes. Next, these hub DEGs were imported into the Kaplan-Meier plotter online database to find the correlation between the DEGs and overall survival (OS) in GC patients. In addition, we also used Gene Expression Profiling Interactive Analysis (GEPIA) to validate the differential expression of DEGs between GC cancer tissues and normal OS tissues. Subsequently, 11 DEGs were found to be eligible, and these were re-analyzed for KEGG pathway enrichment. Finally, three genes [G2/miotic-specific cyclin B1 (CCNB1), polo-like kinases 1 (PLK1), and pituitary tumor-transforming gene-1 (PTTG1)] were identified to be remarkably enriched in the cell cycle pathway, particularly the G1-G1/S phase. Overall, a handful of key genes connected with poor prognosis were discovered via the bioinformatics study, and these may be valid targets for the treatment of GC patients.
We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4427).
Methods
DEGs identification
The GSE33335 and GSE63089 gene expression profiles in GC and normal stomach tissues were acquired from NCBI-GEO (8), which is a free public database of microarray/gene profiles. Microarray data of GSE33335 and GSE63089 separately included 25 GC tissues and 25 matched adjacent noncancerous tissues, along with 45 GC tissues and 45 normal gastric tissues. They were all provided by GPL5175 Platforms. By using GEO2R online tools, the DEGs between GC samples and normal gastric samples were quickly identified. To acquire the co-DEGs among the two datasets, the original data were collated in Venn diagram software. Subsequently, we set|log2(fold change)| >1.2 and false discovery rate (FDR) <0.01 as the cutoff criteria and define selected DEGs as upregulated genes, while the rest were defined as downregulated genes.
GO and pathway enrichment analysis
Defining genes and their products and identifying genome data or genes’ characteristic biological function can be realized by GO analysis. KEGG, a collection of databases, can manage complex information in genomes, and chemical and biological pathways (9). The biological function of DEGs was identified by DAVID (10), an online bioinformatics tool that can be used to visualize the DEG enrichment of biological process (BP), molecular function (MF), cellular component (CC), and pathways.
PPI network and module construction
Search Tool for the Retrieval of Interacting Genes (STRING) is an online tool that can evaluate complex PPI information (11). The potential correlation between these DEGs were examined by the STRING app in Cytoscape (12). And confidence score ≥0.4 and maximum number of interactors = 0 were set as the cutoff criterion. Furthermore, modules of the PPI network were verified by MCODE in Cytoscape with the following standard: degree cutoff =2, max. depth =100, k-core =2, and node score cutoff =0.2.
Survival analysis of hub genes
For estimating the survival rate, we made the use of the Kaplan-Meier plotter, a website tool based on The Cancer Genome Atlas database, the European Genome-phenome Archive (EGA), and Gene Expression Omnibus (GEO) (affymetrix microarrays only) (13). The log-rank P value and hazard ratio (HR) with 95% confidence intervals (CI) were computed and showed on the plot. To validate these DEGs, GEPIA was applied to map the survival plots based on thousands of samples from TCGA (14).
Results
Identification of DEGs and co-DEGs in the two DEGs in GC
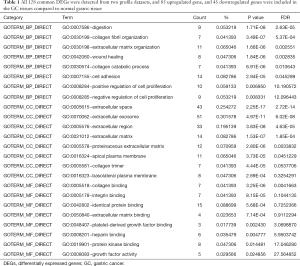
In the present study, 70 GC tissues and 70 normal gastric tissues in DEGs were extracted from GSE33335 and GSE63089 via GEO2R online tools. Then, we identified the common DEGs in the two datasets by using Venn diagram software. A total of 128 common DEGs were detected, including 85 upregulated genes (logFC >1.2 and FDR <0.01) and 43 downregulated genes (logFC <−1.2 and FDR <0.01) in the GC tissues (Table 1, Figure 1).
Full table
GO and KEGG pathway analysis of co-DEGs in GC
We analyzed all 128 co-DEGs in GSE33335 and GSE63089 and completed the GO analysis via DAVID software. The results demonstrated that co-DEGs were mainly enriched in digestion, collagen fibril organization, extracellular matrix (ECM) organization, wound healing, collagen catabolic process, cell adhesion, positive regulation of cell proliferation, negative regulation of cell proliferation (BP); extracellular exosome, extracellular space, extracellular region, ECM, proteinaceous ECM, apical plasma membrane, collagen trimer (CC); identical protein binding, protein kinase binding, collagen binding, integrin binding, heparin-binding, growth factor activity, ECM binding, platelet-derived growth factor binding (MF) (Table 2). For KEGG, the upregulated co-DEGs were mainly involved in the PI3K-Akt signaling pathway, ECM-receptor interaction, protein digestion, absorption, and focal adhesion (Table 3).

Full table
Full table
PPI network construction and modular analysis
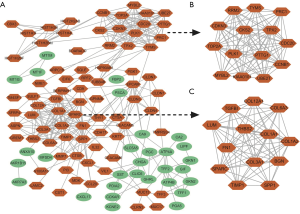
The 128 common DEGs and the top 27 core DEGs with the highest node degree were selected by the network analyzer tool. As presented in Figure 2, the DEG PPI network was constructed using the 128 common DEGs, while the module of the top 27 core DEGs was established by applying Cytotype MCODE (Figure 2B,C).
Analysis of hub genes by the Kaplan-Meier plotter and GEPIA
The survival data of the 27 core genes were calculuated by Kaplan–Meier plotter (http://kmplot.com/analysis) (P<0.05, Figure 3), and 25 genes were associated with significantly worse survival in GC patients via GEPIA (Table 4). Furthermore, 14 of 25 genes were highly expressed in GC samples compared with normal OS samples (P<0.05; Table 5 and Figure 4).
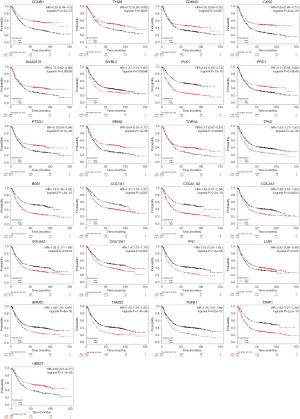

Full table

Full table
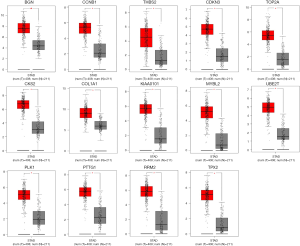
Re-analysis of 14 selected genes via KEGG pathway enrichment
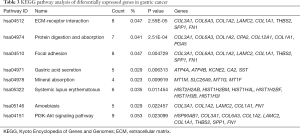
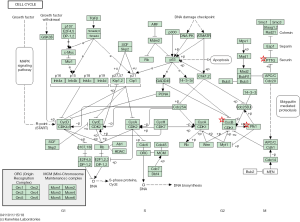
For a better understanding of the possible pathway of these 14 selected DEGs, we used DAVID to re-analyze KEGG pathway enrichment (P<0.05). Three genes (CCNB1, PLK1, and PTTG1) were found to be significantly enriched in the cell cycle pathway, especially in the G1-G1/S phase (Table 6 and Figure 5).

Full table
Discussion
For identifying more useful prognostic biomarkers in GC cancer, bioinformatical methods based on two profile datasets (GSE33335 and GSE63089) were applied in this study, which included 70 GC specimens and 70 normal specimens. A total of 128 commonly changed DEGs (|logFC| >2; adjusted P value <0.05) including 85 upregulated (Log FC >0) and 43 downregulated DEGs were identified (Log FC <0) via GEO2R and Venn diagram software. Next, we used the DAVID online tool to analyse the GO and KEGG pathways of the co-DEGs. The analysis results demonstrated that for KEGG, co-DEGs were particularly enriched in digestion, collagen fibril organization, ECM organization, wound healing, collagen catabolic process, cell adhesion, positive regulation of cell proliferation, negative regulation of cell proliferation (BP); extracellular exosome, extracellular space, extracellular region, ECM, proteinaceous ECM, apical plasma membrane, collagen trimer (CC); in identical protein binding, protein kinase binding, collagen binding, integrin binding, heparin-binding, growth factor activity, ECM binding, platelet-derived growth factor binding (MF). For KEGG, the upregulated co-DEGs were particularly involved in the PI3K-Akt signaling pathway, ECM-receptor interaction, protein digestion, absorption, and focal adhesion.
A DEG PPI network was also constructed, and 27 core genes were screened by Cytotype MCODE analysis. Furthermore, through Kaplan-Meier plotter analysis, 25 of 27 genes were found to be associated with significantly worse survival. In validating these 25 genes with GEPIA analysis, 14 genes showed high expression in GC samples compared with normal samples (P<0.05). Finally, the 14 genes were re-analyzed via DAVID for KEGG pathway enrichment, and then 3 genes (CCNB1, PLK1, and PTTG1) were found to be significantly enriched in the cell cycle pathway, particularly the G1-G1/S phase (P<0.05). These may be regarded as new effective targets for improving the prognosis of GC patients.
CCNB1 is a member of the cyclin B family, and plays a critical role in cells inspecting into or out of M phase in the cell cycle. It is a monitoring protein involved in mitosis and is primarily expressed during the G2/M phase (15). Over the past decades, a large amount of research has demonstrated that CCNB1 is overexpressed in various cancers with poor prognosis, including breast cancer (16), colorectal cancer (17), oral cancer (18), and GC. It was reported that the suppression of CCNB1 by Huang Lian treatment could suppress tumor cell growth in GC by preventing cells from going into the M phase. Moreover, the research by Yasuda et al. shows that the overexpression of CCNB1 occurs in GC and primarily in the early stage. They then further confirmed that high CCNB1 overexpression usually occurs before tumor cells acquire immortalization ability (19). As can be seen, CCNB1 is a well-studied biomarker of GC and is valuable for the prevention and evaluation of therapeutic effects.
PLK1, which belongs to the family of serine/threonine protein kinases, is widespread in eukaryotic cells and has been investigated more intensively than the other four PLKs (20,21). PLK1 takes part in cell mitosis and plays a pivotal role in multiple steps, including G2-M transition, centrosome maturation, bipolar spindle formation, chromosome segregation, DNA replication, and spindle formation (22,23). Decades ago, several studies revealed that the overexpression of PLK1 was closely related to occurrence and the development of malignant tumors (24), including those of GC (25), breast cancer (26), ovarian carcinoma (27), melanoma (28), glioma (29), and renal cancer (30). Furthermore, Wilko et al. demonstrated that (31) PLK1 was overexpressed in roughly half of all gastric carcinomas, and was associated with worse prognosis. Interestingly, in the intestinal metaplasia of normal gastric mucosa, PLK1 has also been found to be expressed or overexpressed. In a relatively new study, we showed that CIP2A, an inhibitor of protein phosphatase 2A, plays a crucial role in facilitating the stability and activity of PLK1 during mitosis by interacting directly with the polo-box domain of PLK1. Thus, the findings above suggest that the CIP2A-Plk1 complex may serve as a potential prognostic marker for poor survival cancer patients. Moreover, small molecules interfering with CIP2A-Plk1 binding could be effective as antimitotic drugs for cancer therapy (32).
PTTG1 is a transcription factor which functions in various physiological events, including transcriptional activity, neovascularization, and cell senescence, and can also participate in cell division, chromosome stability, and DNA repair by encoding regulatory proteins (33,34). After being discovered first in rat pituitary tumor (35), PTTG1 was subsequently reported to be overexpressed in GC (36,37), pituitary adenomas (38), ovarian carcinoma (38), colon carcinoma (39), lung cancer (40), and breast cancer (41). Xu et al. reported that GC tissues expressed a higher level of PTTG1 than adjacent normal tissues. Interestingly, they also found that in GIN, a precancerous lesion, PTTG1 protein expression was significantly higher than para-carcinoma tissues (42,43). Although the specific regulatory mechanisms of PTTG1 in GIN or GC are relatively poorly understood, we can conclude that significant PTTG1 overexpression, both at the mRNA and protein levels, occurring in GC cells in vitro and in vivo, might hold value in GC diagnosis and therapy.
A growing number of studies have implicated these three genes in the emergence and progression of various types of cancers. Unfortunately, few studies have attempted to elaborate the mechanism of action and the precise role of the three genes in GC cancer. We thus hope that the data acquired from our research of GC cancer may offer greater focus to the direction of future studies.
Conclusions
We identified three DEGs (CCNB1, PLK1, and PTTG1) between GC tissues and normal tissues in our bioinformatics analysis study on the base of datasets GSE33335 and GSE63089. Results showed that these three genes could play critical roles in the progression of GC. However, these predictions should be verified by a series of experiments in the future. Overall, this data may provide valuable information and direction for future investigation into the potential biomarkers and biological mechanisms of GC.
Acknowledgments
Funding: This study was funded by the Nantong Science and Technology Project (No. MS12018097).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4427
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4427). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rahman R, Asombang AW, Ibdah JA. Characteristics of gastric cancer in Asia. World J Gastroenterol 2014;20:4483-90. [Crossref] [PubMed]
- Zhang SW, Yang ZX, Zheng RS, et al. Incidence and mortality of stomach cancer in China, 2013. Zhonghua Zhong Liu Za Zhi 2017;39:547-52. [PubMed]
- Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol 2006;12:354-62. [Crossref] [PubMed]
- Fock KM, Ang TL. Epidemiology of Helicobacter pylori infection and gastric cancer in Asia. J Gastroenterol Hepatol 2010;25:479-86. [Crossref] [PubMed]
- Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol 2017;39:1010428317714626. [Crossref] [PubMed]
- Sun Z, Wang Q, Yu X, et al. Risk factors associated with splenic hilar lymph node metastasis in patients with advanced gastric cancer in northwest China. Int J Clin Exp Med 2015;8:21358-64. [PubMed]
- Pritzker KP. Predictive and prognostic cancer biomarkers revisited. Expert Rev Mol Diagn 2015;15:971-4. [Crossref] [PubMed]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2016;44:D7-19. [Crossref] [PubMed]
- Chen L, Zhang YH, Wang S, et al. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One 2017;12:e0184129. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Zhang Y, Lin H, Yang Z, et al. A method for predicting protein complex in dynamic PPI networks. BMC Bioinformatics 2016;17 Suppl 7:229. [Crossref] [PubMed]
- Doncheva NT, Morris JH, Gorodkin J, et al. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J Proteome Res 2019;18:623-32. [Crossref] [PubMed]
- Ren J, Niu G, Wang X, et al. Overexpression of FNDC1 in Gastric Cancer and its Prognostic Significance. J Cancer 2018;9:4586-95. [Crossref] [PubMed]
- Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Lin Z, Tan C, Qiu Q, et al. Ubiquitin-specific protease 22 is a deubiquitinase of CCNB1. Cell Discov 2015;1:15028. [Crossref] [PubMed]
- Ding K, Li W, Zou Z, et al. CCNB1 is a prognostic biomarker for ER+ breast cancer. Med Hypotheses 2014;83:359-64. [Crossref] [PubMed]
- Fang Y, Yu H, Liang X, et al. Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biol Ther 2014;15:1268-79. [Crossref] [PubMed]
- Patil GB, Hallikeri KS, Balappanavar AY, et al. Cyclin B1 overexpression in conventional oral squamous cell carcinoma and verrucous carcinoma- A correlation with clinicopathological features. Med Oral Patol Oral Cir Bucal 2013;18:e585-90. [Crossref] [PubMed]
- Yasuda M, Takesue F, Inutsuka S, et al. Overexpression of cyclin B1 in gastric cancer and its clinicopathological significance: an immunohistological study. J Cancer Res Clin Oncol 2002;128:412-6. [Crossref] [PubMed]
- Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer 2010;10:825-41. [Crossref] [PubMed]
- de Carcer G, Escobar B, Higuero AM, et al. Plk5, a polo box domain-only protein with specific roles in neuron differentiation and glioblastoma suppression. Mol Cell Biol 2011;31:1225-39. [Crossref] [PubMed]
- Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol 2004;5:429-40. [Crossref] [PubMed]
- Ando K, Ozaki T, Yamamoto H, et al. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem 2004;279:25549-61. [Crossref] [PubMed]
- Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene 2005;24:267-76. [Crossref] [PubMed]
- Erratum: PLK1 as a potential prognostic marker of gastric cancer through MEK-ER K pathway on PDTX models Onco Targets Ther 2019;12:413. [Corrigendum]. [Crossref] [PubMed]
- Weichert W, Kristiansen G, Winzer KJ, et al. Polo-like kinase isoforms in breast cancer: expression patterns and prognostic implications. Virchows Arch 2005;446:442-50. [Crossref] [PubMed]
- Weichert W, Denkert C, Schmidt M, et al. Polo-like kinase isoform expression is a prognostic factor in ovarian carcinoma. Br J Cancer 2004;90:815-21. [Crossref] [PubMed]
- Kneisel L, Strebhardt K, Bernd A, et al. Expression of polo-like kinase (PLK1) in thin melanomas: a novel marker of metastatic disease. J Cutan Pathol 2002;29:354-8. [Crossref] [PubMed]
- Cheng MW, Wang BC, Weng ZQ, et al. Clinicopathological significance of Polo-like kinase 1 (PLK1) expression in human malignant glioma. Acta Histochem 2012;114:503-9. [Crossref] [PubMed]
- Zhang G, Zhang Z, Liu Z. Polo-like kinase 1 is overexpressed in renal cancer and participates in the proliferation and invasion of renal cancer cells. Tumour Biol 2013;34:1887-94. [Crossref] [PubMed]
- Weichert W, Ullrich A, Schmidt M, et al. Expression patterns of polo-like kinase 1 in human gastric cancer. Cancer Sci 2006;97:271-6. [Crossref] [PubMed]
- Kim JS, Kim EJ, Oh JS, et al. CIP2A modulates cell-cycle progression in human cancer cells by regulating the stability and activity of Plk1. Cancer Res 2013;73:6667-78. [Crossref] [PubMed]
- Malik MT, Kakar SS. Regulation of angiogenesis and invasion by human Pituitary tumor transforming gene (PTTG) through increased expression and secretion of matrix metalloproteinase-2 (MMP-2). Mol Cancer 2006;5:61. [Crossref] [PubMed]
- Salehi F, Kovacs K, Scheithauer BW, et al. Pituitary tumor-transforming gene in endocrine and other neoplasms: a review and update. Endocr Relat Cancer 2008;15:721-43. [Crossref] [PubMed]
- Pei L, Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG). Mol Endocrinol 1997;11:433-41. [Crossref] [PubMed]
- Luo Z, Li B, Chen J, et al. Expression and the clinical significance of hPTTG1 in gastric cancer. Mol Med Rep 2013;7:43-6. [Crossref] [PubMed]
- Wen CY, Nakayama T, Wang AP, et al. Expression of pituitary tumor transforming gene in human gastric carcinoma. World J Gastroenterol 2004;10:481-3. [Crossref] [PubMed]
- Zhang X, Horwitz GA, Heaney AP, et al. Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. J Clin Endocrinol Metab 1999;84:761-7. [Crossref] [PubMed]
- Panguluri SK, Yeakel C, Kakar SS. PTTG: an important target gene for ovarian cancer therapy. J Ovarian Res 2008;1:6. [Crossref] [PubMed]
- Heaney AP, Singson R, McCabe CJ, et al. Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet 2000;355:716-9. [Crossref] [PubMed]
- Rehfeld N, Geddert H, Atamna A, et al. The influence of the pituitary tumor transforming gene-1 (PTTG-1) on survival of patients with small cell lung cancer and non-small cell lung cancer. J Carcinog 2006;5:4. [Crossref] [PubMed]
- Ghayad SE, Vendrell JA, Bieche I, et al. Identification of TACC1, NOV, and PTTG1 as new candidate genes associated with endocrine therapy resistance in breast cancer. J Mol Endocrinol 2009;42:87-103. [Crossref] [PubMed]
- Xu MD, Dong L, Qi P, et al. Pituitary tumor-transforming gene-1 serves as an independent prognostic biomarker for gastric cancer. Gastric Cancer 2016;19:107-15. [Crossref] [PubMed]
(English Language Editor: J. Gray)


