
Current practices on genetic testing in ovarian cancer
Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecological cancer. It represents the seventh most commonly diagnosed cancer among women in the world with a 5-year survival rate of 46%. Annually worldwide, 295,000 women will be diagnosed and 184,000 will die. In 2018 in the United States, there were approximately 22,240 new cases of ovarian cancer diagnosed and 14,070 ovarian cancer deaths (1-3). One of the main factors contributing to the high death-to-incidence rate is the advanced stage of the disease at the time of diagnosis. Therefore, improving prevention and early detection is a research priority because disease diagnosed at an early stage has a 5-year relative survival rate that exceeds 90% (4,5). EOC encompasses a heterogenous group of malignancies that vary in etiology, molecular biology, and numerous other characteristics and is divided according to histologic subtypes: high-grade serous (the most frequent), low-grade serous, clear cell, endometrioid, and mucinous (6). Each histologic subtype is associated with a distinct clinical behavior (response to chemotherapy, pattern of metastases, survival) but has historically been treated as one entity (6-9).
The majority of hereditary ovarian cancer can be attributed to germline BRCA1 & BRCA2 pathogenic variants. A number of additional variants, in genes beyond BRCA1/2, have been identified and are suspected to play a significant role in ovarian carcinogenesis. As the complexity of genetic testing increases, the evolution of next generation sequencing (NGS) allows rapid evaluation of variants in multiple cancer susceptibility genes at similar costs to single gene sequencing.
Molecular landscape
Homologous recombination (HR) repair pathway
DNA double-strand breaks (DSBs) are one of the most common and most cytotoxic types of DNA damage that can consequently result in significant genomic aberrations, which if left unrepaired or improperly repaired, will ultimately result in cell death (10). In humans, DSBs are repaired by a number of repair pathways, the most important of which involve non-homologous end joining (NHEJ) and HR (11). Of these, HR is the repair mechanism which is performed with high fidelity.
The process of HR repair, in which a number of proteins are involved, starts with DSBs recognition by the MRN complex that consists of three proteins: MRE11, RAD50, and NBS1. During the initial recruitment, MRN complex binds through its MRE11 component to DSB. Subsequently, RAD50 arms undergo structural changes, thereby allowing the bridging of both DNA ends. In a next step, signaling cascade is activated with the activation of the ATM kinase. The ATM kinase phosphorylates many other DNA damage response proteins, including members of the MRN complex (12).
One crucial step in HR repair is the formation of single-strand DNA overhangs at the sites of DNA ends. This process requires resection of 5' termini to generate 3' single-strand DNA overhangs (tails) and is initiated by the MRN complex in conjunction with CtIP and BRCA1 (10,13,14). More specifically, the bifunctional endo/exonuclease MRE11 can initiate DNA-end resection in 5'-3' direction and begins by removing ~200–300 nucleotides away from the DSB site (15). This initial resection results in relatively short single-strand DNA overhangs and is followed by mass resection of DNA by either the 5'-3' exonuclease 1 or DNA 2 in conjunction with the helicase Sgs1, thereby generating extensive single-strand DNA stretches (11,14,15). Subsequently, single-strand DNA is coated with replication protein A (RPA) complexes. In parallel, BRCA2 is recruited in a BRCA1- and PALB2-dependent manner to ultimately recruit recombinase RAD51, a DNA-dependent ATPase, to the single-strand DNA overhangs. Loading of RAD51 into single-strand DNA is a key step in HR repair. RAD51 replaces RPA and forms nucleoprotein filaments in single-strand DNA, which are essential for the homology search in sister chromatid and strand exchange (10,15,16).
Intermediary metabolic products of streptomycetes, including bleomycin, neocarzinostatin, and related compounds are effective cytotoxic agents. They directly induce DSB by attacking specific carbons in deoxyribose, leaving non-standard end-groups (17-19). Furthermore, topoisomerases inhibitors have DNA-based anti-tumor efficacy. Topoisomerases types I and II are enzymes that open and close one and both strands of DNA, respectively. Both types involve a covalent DNA-protein bond in their catalytic cycle (20,21). This transition state can be stabilized by topoisomerase inhibitors leading to DSB for type II inhibitors (epipodophyllotoxins) or single strand break (SSB) for type I inhibitors (camptothecins). Topoisomerases are covalently attached to the 5'-ends (bacterial type I, bacterial and eukaryotic type II) or 3'-ends (eukaryotic type I) of the broken DNA (22,23).
Poly(ADP-ribose) polymerase (PARP): mechanism of action
The PARPs are a family of enzymes capable of catalyzing the transfer of ADP-ribose to target proteins. To date, 17 members of the PARP family have been identified. PARP-1 is the first and best characterized member and accounts for ~90% of the total PARP activity. It plays a main role in DNA base excision repair (BER) and repair of DNA SSBs. Human PARP-1 has a multi-domain architecture composed of six domains. PARP-1 binding to the ends of an SSB is achieved through the coordinated action of two zinc finger domains located at the N-terminus of the protein. The catalytic domain is composed of an ADP-ribosyl transferase fold and a helical subdomain (10,24,25). PARP-1 binding to SSBs is followed by the polymerization of ADP-ribose (PARylation) with NAD+ as a substrate, resulting in the production of variable-sized polymers of ADP-ribose (PAR) (11). Successive addition of ADP-ribose units to form long and branched poly(ADP-ribose) chains, covalently linked to receptor proteins, including PARP-1, histone and other DNA repair proteins, leads to the formation of polymers adjacent to the DNA breaks. These highly negatively charged polymers form a scaffold and recruit other proteins, such as XRCC1, DNA ligase 3 and DNA polymerase β, that are critical in SSB repair through the BER pathway. If PARylation is inhibited and BER is impaired, unrepaired SSBs accumulate, and they degenerate in replication forks to become DSBs (10,11).
Dawson and coworkers first connected PARP-1 hyperactivation with apoptosis-inducing factor (AIF) release from mitochondria (26). The upregulated PAR synthesis resulting from PARP-1 overactivation initiates a nuclear signal that propagates to mitochondria and triggers the release of AIF, which activates caspase-independent cell death (27).
Synthetic lethality
Synthetic lethality was initially described in 1922 by Calvin Bridges, an American geneticist who helped establish the chromosomal basis of heredity and sex. He found that certain non-allelic genes were lethal only in combination, during crossover between Drosophila melanogasters (28). Indeed, the term describes the lethal effect of two simultaneous genetic aberrations, which when occurring in isolation are otherwise non-lethal (29,30). This phenomenon has attracted attention due to the prospect of designing novel drugs and was described by two independent groups in 2005, when PARP inhibitors (PARPis) showed efficacy against HR-defective cell lines and tumor xenografts or allografts. More specifically, Bryant et al. (31) described the profound cytotoxicity, when even low concentrations of two PARPis were used in HR-defective cells (including human breast cancer cells). In addition, Farmer et al. (32) reported the vulnerability of cells that were BRCA1 or BRCA2 deficient to the inhibition of PARP enzymatic activity, which subsequently resulted in chromosomal instability, cell cycle arrest and ultimately, apoptosis. The concept of synthetic lethality of PARPi in HR defective cells is generally believed to be due to SSBs repair failure. Such SSBs will result in collapsed replication forks and replication-associated DSBs that require HR system for repair. In the absence of HR, these lesions prove lethal either because they persist or they can only be repaired by alternative pathways, such as NHEJ, that are prone to errors (25,29,30).
Mismatch repair (MMR)
The MMR system, an evolutionarily conserved mechanism, plays an important role in maintaining genomic stability. It recognizes and corrects biosynthetic errors that arise during DNA replication as well as mispaired bases that are generated during recombination or caused by oxidative DNA damage. Therefore, MMR reduces DNA errors 100–1,000 fold, and prevents them from becoming fixed mutations during cellular proliferation (33-37). Seven MMR genes, MLH1, MLH3, MSH2, MSH3, MSH6, PMS1, and PMS2 are involved in human MMR function.
Hereditary nonpolyposis colorectal cancer (HNPCC) or Lynch syndrome is caused by mutations in the genes that are responsible for MMR and is associated with a high risk of colon cancer, as well as other cancers including endometrial cancer, ovary, stomach, hepatobiliary tract, small intestine, upper urinary tract, brain, and skin. In ovarian cancer, MMR deficiency is the most common cause of hereditary ovarian cancer after HR deficiency accounting for another 10–15% of hereditary ovarian carcinomas (36,38). Pooled data from four large hereditary cancer registries in Europe and the United States determined a 6.7% lifetime risk for ovarian cancer in proven or probable MSH2 and MLH1 mutation carriers from Lynch syndrome families. Overall, women with HNPCC syndrome have approximately a 12–15% lifetime risk of developing ovarian cancer (39). In addition to the inherited gene mutations, other mechanisms of gene inactivation (such as promoter hypermethylation) leading to loss of expression of one of the main MMR genes occurs in up to 29% of cases (36).
Furthermore, there is published evidence that high mRNA levels of MSH6, MLH1, and PMS2 were associated with a favorable overall survival (OS) in ovarian cancer, suggesting these MMR genes may serve as potential positive prognostic indicators in ovarian cancer patients treated with platinum-based chemotherapy (40).
Susceptibility to EOC
Prevalence of pathogenic sequence variants (PVs) in cancer predisposing genes
Ovarian cancer is probably the tumor type with the highest percentage of hereditary cases observed, irrespectively of selection criteria, such as early age at onset or family history. More specifically, a fourth to a fifth of unselected EOC patients carry PVs in a number of genes, the majority of which, encode for proteins involved in DNA repair pathways (41). BRCA1 and BRCA2 predisposing variants were the first to be associated to ovarian cancer, with the advent in DNA sequencing technologies leading to the discovery and association of additional genes. PVs of many of the genes which encode for proteins involved in HR have been associated with increased risk of ovarian cancer, due to the significant role of HR in ovarian carcinogenesis (1,42-44).
The Cancer Genome Atlas (TCGA) found HR to be defective in approximately half of 489 women with stage II to IV high-grade serous ovarian cancers (HGSOCs) (45) attributed to germline variants in BRCA1 (in 8.5% of tumors) or BRCA2 (6.3%), somatic variants in BRCA1 (3.2%) or BRCA2 (2.9%) or epigenetic inactivation of BRCA1 (10.8%) due to promoter methylation resulting in transcriptional silencing. The prevalence of epigenetic/genetic mechanisms of BRCA1 aberration have been noted to vary between ethnicities, with PVs predominating in White Europeans, and methylation in people of African descent (46). Alsop et al. reported germline BRCA1 or BRCA2 PVs in approximately 15% of EOC patients, and 23% of patients with HGSOC (47).
In addition to the most commonly occurring alterations in BRCA1/2 genes, genetic aberrations in other genes compromise the HR pathway, including the Fanconi anemia genes (PALB2, PRIB1), the core RAD genes (RAD51C, RAD51D) and genes involved in HR pathway either directly (CHEK2, BARD1, NBN, ATM) or indirectly (such as CDK12, that encodes a cyclin-dependent kinase involved in the transcription regulation of several HR genes) (48-51). PVs in many of these genes confer increased, but variable, risks for ovarian cancer development. In addition, as previously mentioned, PVs genes involved in MMR pathway, account for 10–15% of hereditary EOC and is typically associated with tumors of endometrioid or clear-cell histopathology (1). Furthermore, PTEN downregulation (6.7%) has been associated with RAD51 transcriptional downregulation, while EMSY overexpression and amplification (7.9%) has been reported as another mechanism of HR deficiency in women with HGSOCs (1,50-52).
The majority of the aforementioned genes are included and analyzed in multi-gene panels utilized for hereditary cancer predisposition. HR deficient EOC has greater sensitivity to DNA-damaging agents that crosslink DNA, such as cisplatin, because HR is required for the repair of these lesions. Therefore, the identification of women with HR deficient ovarian cancers has significant clinical implications concerning chemotherapy regimen planning and development and use of targeted therapies as well. Cascade testing should also be offered to blood relatives of PV carriers.
Germline predisposing variants (Figure 1)
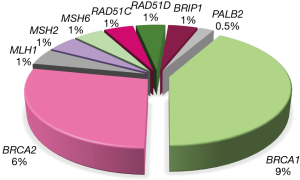
BRCA1 and BRCA2
PVs in BRCA1 and BRCA2 genes are detected in 10–15% (47,53-55) of unselected EOC patients, accounting for the majority of hereditary cases. BRCA1 and BRCA2 PVs confer 44% and 17% lifetime risks for ovarian cancer diagnosis, respectively (56), in contrast to ~1% risk for diagnosis in the general population. The vast majority of women carrying BRCA1 and BRCA2 PVs will be diagnosed with ovarian cancer, specifically with high-grade serous histology, with a median age of onset at ~51 and ~61 years, respectively (41,53,57). On the contrary, the largest prospective study performed to date, showed that the risk for ovarian cancer diagnoses was low up to the age of 40 and 50 years for BRCA1 and BRCA2 carriers, respectively (56). Ovarian cancers that bare pathogenic BRCA1 & BRCA2 variants are characterized by distinct clinical behaviors, illustrated by improved survival, visceral disease distribution, higher response rates to platinum and certain non-platinum chemotherapy agents, and sensitivity to PARPis.
Identification of BRCA1 and BRCA2 PVs can have major impact on appropriate clinical management of cancer patients through the choice of tailored treatments. Beyond the proven clinical benefit observed on the patients themselves, the genetic information can be used as a measure of primary prevention for ovarian cancer and is therefore critical for their family relatives. Asymptomatic carriers, that can be identified through cascade testing, and are at high risk, can be offered risk-reducing options that can ultimately lead to reduced morbidity and mortality. Simultaneously, through testing, all family relatives that will test negative, can be assured that they have a lifetime ovarian risk, similar to the general population.
The proposed primary prevention measure for ovarian cancer risk reduction among BRCA1 and BRCA2 carriers is bilateral salpingo-oophorectomy (BSO), as regular surveillance has been shown to be inadequate to detect tumors at an early stage. BSO, which can reduce the risk for ovarian cancer diagnosis up to 96% (57,58), should be considered after extensive counselling and discussion with the individual and of course, after completion of childbearing. Ideally, for BRCA1 carriers the procedure should be performed after the age of 35 and before the age of 40 years. For BRCA2 carriers, taking into consideration the lower penetrance and later onset of diagnosis, BSO can be performed after the age of 40 and before the age of 45 years (59,60). Keeping this age limits for BSO, the vast majority of ovarian cancers arising in the context of BRCA1 and BRCA2 predisposing variants will be avoided.
RAD51C and RAD51D
One of the co-localizing molecules with BRCA1 and BRCA2 in the repair of double-strand DNA breaks is RAD51C. RAD51C loss-of-function variants are rare among ovarian cancer patients, with their prevalence varying between 0.3–1.1% (61), with the upper end corresponding to studies enriched with cases with breast and ovarian cancer family history. The lifetime risk for ovarian cancer diagnosis among RAD51C carriers is ~5% (51,61). PVs in an additional member of the RAD51 family are also associated with increased ovarian cancer risk and are more frequently identified in familial ovarian cancer cases, reaching a prevalence of 5.1% in families with at least three ovarian cancer cases (62). Damaging RAD51D variants are marginally less frequent among ovarian cancer patients, compared to RAD51C PVs, observed in ~0.2% of unselected ovarian cancer patients (61,63) up to 0.9% of individuals having strong family history (62). RAD51D loss-of-function variants are associated with at least a six-fold increased risk for ovarian cancer, therefore identification of such genetic defects can have clinical utility (61,62).
Both RAD51C and RAD51D are ovarian cancer susceptibility genes, showing a reduced magnitude, when compared to BRCA1 and BRCA2, while identification of causative variants in females can lead informed decisions on preventative measures. PVs in any of these genes confer sufficient risk to guide risk-reducing BSO, as proposed in the National Comprehensive Cancer Network (NCCN) guidelines (64). Furthermore, it is plausible that genetic defects in these genes can facilitate as biomarkers for PARP sensitivity. Interestingly, there have been developed decision analytic models for the identification of the ovarian cancer risk thresholds, appropriate for risk reducing salpingo-oophorectomy-based primary prevention. Cost-effectiveness analysis has been highlighted by the National Institute of Health and Care Excellence (NICE) as the favored form of economic assessment to compare relative costs and health outcomes in decision modeling (65).
BRIP1
Heterozygous BRIP1 (BRCA1-interacting protein C-terminal helicase-1) damaging variants are associated to ovarian cancer susceptibility. The prevalence of PVs among familial ovarian cancer patients can be as high as 2.8% (66) or ~1% among unselected patients (51,61). The cumulative lifetime risk for EOC diagnosis among BRIP1 carriers has been estimated as 5–5.8% (61,67), with the most cases being diagnosed after menopause. Among the studies reporting the largest series of BRIP1 carriers, the mean age at diagnosis was 61–64 years (51,66,67). The elevated risk for EOC diagnosis justifies recommendation for risk-reducing removal of salpinges and ovaries among asymptomatic carriers and should be guided based on family history and individual’s choice, which can be offered at a later age, when compared to BRCA1 and BRCA2 carriers.
PALB2
Clinical testing for PALB2 (partner and localizer of BRCA2) has become a standard of practice, when evaluating breast cancer predisposition, while encoding for a key protein that works closely with BRCA1 and BRCA2 to facilitate DNA repair through HR. Nevertheless, the association of PALB2 PVs to ovarian cancer susceptibility is still conflicting, with the majority of studies reporting relative risks that ranged from 0.9 to 5.5 and lacked statistical power (52,67,68). Through monitoring 524 families known to carry PALB2 PVs, the risk to EOC predisposition was estimated to reach 5% by the age of 80 years (69). For PALB2 carriers, discussion and recommendation for risk-reducing surgery should take place for cases with strong family history for ovarian cancer. It should also be taken under consideration, that PALB2 PVs are indeed quite rare among ovarian cancer patients, identified in less than 0.5% of cases interrogated (51,61,67). A quite important observation, which provides promising therapeutic interventions, involves the sensitivity of PALB2 associated tumors to platinum-based chemotherapy and PARPis (46,70).
MLH1, MSH2, MSH6 and PMS2
Loss-of-function variants in the MMR genes, MLH1, MSH2, MSH6 and PMS2, predispose for HNPCC syndrome. Although the main phenotypic hallmarks in individuals with HNPCC syndrome involve colorectal and endometrial cancer diagnoses, the cancer spectra are relatively broad, as previously mentioned. Approximately 1% of all EOC cases can be attributed to PVs in MMR genes (71). Cancer risk among HNPCC syndrome individuals is gene-specific, i.e., damaging variants in each of the MMR genes confer variable risks. MLH1 and MSH2 PVs are associated with higher penetrance of both colorectal and endometrial cancer diagnoses, both estimated to be more than 50% to the age of 70 years (39,72-74), while showing phenotypic variability in terms of organ sites. MSH2 PVs seem to be associated with the higher risk for ovarian cancer diagnoses, conferring a risk of 10–24%, followed by MLH1 PVs, which are reported to be associated with a risk of 5–20%, by the age of 70 years (73,74). MSH6 PVs confer much lower risks, ranging from 1–11%, while PMS2 PVs seem not to be associated with elevated ovarian cancer risk (39,71,74). The majority of ovarian cancer in the context of HNPCC syndrome will occur before the age of 50 years with the age range being 43 to 44 years, 44 to 47 years and 44 to 48 years for MSH2, MLH1 and MSH2 carriers, respectively (71-75).
A systematic review on ovarian cancer occurring in the context of HNPCC syndrome showed that the most prominent histological type was the high-grade endometrioid adenocarcinoma, either diagnosed solely or along with other types, accounting for more than half of the cases. Moreover, high-grade serous subtype, accounts for ~20% of the cases, while both clear-cell and mucinous histology can occur (76,77). Immunohistochemistry for protein expression of MMR and/or testing for microsatellite instability (MSI) of the pathologic specimen of the ovarian tumor can be used as a screening tool, although it is not widely incorporated or tested. Based on NCCN guidelines (64), women with HNPCC syndrome (mostly referring to women carrying MLH1 and MSH2 PVs) should consider risk-reducing surgery, after childbearing, which will involve hysterectomy and BSO, in order to significantly reduce overall gynecological cancer occurrence. Such a decision should be taken following genetic counselling sessions, where all the benefits and potential risks of such a procedure have been discussed. The optimal timing for prophylactic surgery can vary and depends on multiple factors involving family history, age at diagnosis of gynecological cancers among family relatives, the MMR gene where the PV is identified and the age of childbearing completion. Irrespectively of the time point when the surgery will be performed tough, all these women should be under close surveillance by annual transvaginal ultrasound, random endometrial biopsies every 1–2 years and measurement of serum levels of CA125 on a year basis. Of course, the aforementioned means of surveillance have not been proven as an efficient method of prophylaxis and an important aspect of primary diagnosis is indefinitely, women education on possible symptoms that might occur (39).
Tumor testing in ovarian cancer
The need to increase the number of ovarian cancer patients that can potentially benefit from treatment with targeted therapies, such PARPis, has significantly increased the need for tumor testing through which additional genetic changes, which can predict sensitivity to PARP inhibition, can be identified. Indeed, an additional 4–7% of BRCA1 and BRCA2 PVs can be identified through tumor testing in EOC patients previously tested negative for germline loss-of-function BRCA1 and BRCA2 variants, while ~3% can bear somatic variants in other genes that are implicated in HR (78,79).
Hypothetically, through tumor testing, both germline and somatic genetic alterations can be detected, whereas germline testing alone cannot identify somatic mutations. Although the trend for tumor testing alone seems appealing, there are a number of factors that can influence the accuracy of such testing, while it can’t be considered as a gold standard for germline variant detection. Most importantly, in most cases the starting material for tumor testing is DNA extracted from formalin-fixed paraffin-embedded (FFPE) tissues, which can be technically challenging to amplify and to provide accurate results. In order to obtain tumor DNA, tumor microdissection is required. This means that in practical terms, a small diagnostic biopsy is likely to be unsuitable for further analysis. Of note, poor fixation can result in fragmented DNA; while the formalin itself can induce C>U deamination, leading to sequencing errors and the risk of false mutation calls. Another important aspect that should be taken into account is tumor heterogeneity and the actual percentage of tumor cells that are included in the tumor specimen from which DNA to be analyzed, will be extracted. Lastly, but importantly, one should take into account the limitations that these exhibit. Detection of large genomic rearrangements or PVs in GC genomic areas can be quite challenging in FFPE DNA (80), while the phenomenon of genetic reversion of the initial BRCA1 or BRCA2 variant that can occur most frequently as a result of chemotherapy, or even as an adaptive resistance mechanism (81). Given these considerations, although tumor testing can be proven of high importance during therapeutic decision-making, it certainly cannot replace traditional germline testing for assessing predisposition to hereditary cancer.
Testing for HR deficiency
It is evident that the majority of ovarian tumors presented with HR deficiency will derive from individuals harboring germline PVs in BRCA1 and BRCA2. However, a number of tumors can have impaired HR pathway, as a result of alternative genetic defects, occurring either in the germline or in the soma, as previously described. Indeed, based on the data from TCGA, as much as half of the high-grade ovarian cancers harbor PVs in genes involved in the HR pathway (45). Of course, it has to be clarified that not all genetic alterations in these genes will result in HR deficiency.
Identification of HR deficiency in a tumor can be reflected through additional assays which can determine the genomic instability and can provide an HR deficiency score by measuring: the generic loss of heterozygosity (LOH), telomeric allelic imbalance and large-scale state transitions. Such functional assays aim to determine the genomic scars that are created following accumulation of genomic alterations and have been mainly developed and patented by commercial companies. Both LOH and HR deficiency score have been implemented as biomarkers, by a number of trials testing the efficiency of different PARPis, such as niraparib (82) and rucaparib (83). Interestingly, in the latter ARIEL2 study, 34% of BRCA wild type patients with LOH low archival specimen had an LOH high pretreatment specimen highlighting a change in biomarker status over time. In contrast, there was no case in which the classification changed from LOH high to LOH low between the archival and pretreatment specimen. Based on the variability and the complexity of the different genetic phenomena that can occur in an ovarian tumor, the available tests should provide higher levels of accuracy when ordered and used in a complementary manner.
The role of PARPis in EOC and their therapeutic potential in patients with HR deficiency (Table 1)
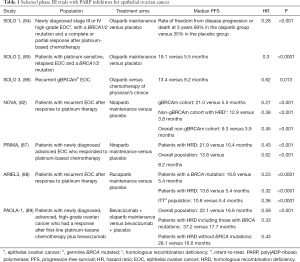
Full table
PARPis have transformed treatment for ovarian cancer. Olaparib received FDA approval based on the results from Study 19. This phase II study randomized women with relapsed platinum-sensitive HGSOC who had received at least two prior platinum-based chemotherapy regimens and were in complete or partial response to their most recent regimen to olaparib maintenance treatment or placebo. Olaparib treatment improved progression-free survival (PFS) in the overall population (hazard ratio 0.35, P<0.0001). In the BRCA mutant population, the benefit of olaparib versus placebo was even greater than in the BRCA wild-type population (90). The SOLO2 phase III trial was designed to confirm the efficacy of olaparib maintenance in patients with BRCA mutated, relapsed EOC who were in complete or partial response to their most recent platinum-based regimen. Olaparib maintenance treatment provided a significant PFS improvement with no detrimental effect on quality of life in these patients (85). Another phase III trial (SOLO1), evaluated the efficacy of olaparib as maintenance therapy in patients with newly diagnosed advanced EOC with a mutation in BRCA1, BRCA2, or both who had a complete or partial clinical response after first-line platinum-based chemotherapy. The risk of disease progression or death was 70% lower with olaparib than with placebo (84). Finally, in the phase III SOLO3 trial patients with platinum sensitive, relapsed EOC with germline BRCA mutations were randomized to olaparib or non-platinum chemotherapy of physician’s choice. The study demonstrated that treatment with the PARPi resulted in better outcomes [overall response rate (ORR) and PFS] than non-platinum chemotherapy in these women (86).
Trials with two other PARPi, niraparib and rucaparib, have confirmed the efficacy of these drugs as maintenance therapy. More specifically, the phase III ENGOT-OV16/NOVA trial evaluated the efficacy of niraparib versus placebo as maintenance treatment for patients with platinum-sensitive, recurrent ovarian cancer. Patients in the niraparib group had a significantly longer median PFS than did those in the placebo group (21.0 versus 5.5 months in the germline BRCA mutant cohort, 12.9 versus 3.8 months in the non-germline BRCA mutant cohort with HR deficiency, and 9.3 versus 3.9 months in the overall non-germline BRCA mutant cohort) (82). Niraparib was also evaluated in women with newly diagnosed advanced EOC who had a response to platinum-based chemotherapy (PRIMA/ENGOT-OV26/GOG-3012 trial). Among 373 (50.9%) patients with HR deficiency, the median PFS was 21.9 months in the niraparib group and 10.4 months in the placebo group (hazard ratio 0.43, P<0.001). In the entire population, the corresponding PFS was 13.8 and 8.2 months, respectively (hazard ratio 0.62, P<0.001). At 24 months, the rate of overall survival was 84% in the niraparib group and 77% in the placebo group (87).
The phase III ARIEL3 trial, assessed rucaparib versus placebo after response to second-line or later platinum-based chemotherapy in patients with high-grade, recurrent, platinum-sensitive ovarian carcinoma. Median PFS in patients with a BRCA-mutant carcinoma (germline or somatic) was 16.6 months in the rucaparib group versus 5.4 months (hazard ratio 0.23, P<0.0001). In patients with a HR deficient carcinoma it was 13.6 versus 5.4 months (hazard ratio 0.32, P<0.0001), while in the intention-to-treat population, it was 10.8 versus 5.4 months (hazard ratio 0.36, P<0.0001). Thus, ARIEL3 provided further evidence that use of a PARPi in the maintenance treatment setting could be considered a new standard of care for women with platinum-sensitive EOC following a complete or partial response to second-line or later platinum-based chemotherapy (88). These trials clearly indicate that the relationship between sensitivity to PARPis and HR repair deficiency is likely to be more similar to a continuous than to a fixed and discrete variable. As such, PARPis have the potential to be beneficial in a much wider proportion of EOC patients than was originally proposed (91).
More recently, because PARP inhibition has immuno-regulatory effects (92,93), combination therapy of PARP and immune checkpoint inhibitors is being developed. A phase II study (MEDIOLA) of olaparib and the PD-L1 inhibitor durvalumab in patients with relapsed, platinum-sensitive, BRCA-mutated ovarian cancer showed an ORR of 72% (94). The phase I/II study TOPACIO investigated the combination of niraparib and pembrolizumab in patients with platinum-resistant/refractory ovarian cancer (95). The ORR was 18% and the disease control rate was 65%. There was no difference in response by BRCA and homologous recombination deficiency (HRD) status. Currently, several studies that combine PARPis and immune check point inhibitors are being carried out. Among them, two interesting clinical trials should be mentioned: ENGOT-OV44/FIRST study compares the efficacy and safety of standard of care platinum-based therapy ± dostarlimab (an anti-PD-1 humanized monoclonal antibody that has shown clinical activity as monotherapy in early phase trials) followed by niraparib ± dostarlimab maintenance as first-line treatment of stage III or IV EOC (96); ATHENA (GOG-3020/ENGOT-OV45) is a randomized, double-blind, placebo-controlled, phase 3 study of rucaparib + nivolumab following frontline platinum-based chemotherapy (97).
Because PARPis and anti-angiogenic inhibitors are also synergistic (92,98), the combination of these drugs also has been studied. In the phase 3 trial PAOLA-1, patients with newly diagnosed, advanced, high-grade EOC having a response after first-line platinum-taxane chemotherapy plus bevacizumab were randomly assigned in a 2:1 ratio to receive olaparib or placebo for up to 24 months and bevacizumab for up to 15 months in total. Maintenance with olaparib provided a significant PFS benefit, which was substantial in patients with HRD-positive tumors, including those without a BRCA mutation (89).
Beyond PAOLA-1, several other trials are examining the role of anti-angiogenesis, in combination with PARPis in the different treatment lines of EOC, and different subpopulations as well. In most studies, the combination of cediranib [an oral, highly potent, VEGF receptor (VEGFR) tyrosine kinase inhibitor with activity against all three VEGFRs] with olaparib is tested. The randomized, phase II AVANOVA2 trial compared niraparib and bevacizumab versus niraparib alone as definitive treatment for platinum-sensitive recurrent ovarian cancer (99). Niraparib plus bevacizumab significantly improved PFS compared with niraparib alone (median PFS 11.9 versus 5.5 months, respectively; adjusted hazard ratio 0.35, P<0.0001). Following the encouraging results of a randomized phase II trial (NCT01116648) which showed significant improvement in PFS with cediranib + olaparib versus olaparib monotherapy in recurrent platinum-sensitive EOC, notably in non-germline BRCA mutated patients (100), the phase IIb CONCERTO study was conducted. This study investigated the combination of cediranib plus olaparib in non-germline BRCA mutated patients with recurrent platinum-resistant EOC who had received ≥3 previous lines of therapy for advanced disease. The combination showed evidence of antitumor activity in this heavily pretreated subpopulation, while toxicity was manageable with dose modifications (100). In contrast to the previous study, the randomized, phase II OCTOVA trial, investigates the benefit of single agent olaparib compared to olaparib plus cediranib or weekly paclitaxel in women with BRCA-mutated platinum-resistant EOC (101). Also, a randomized phase II/III study (COCOS), compares of the combination of cediranib and olaparib to cediranib or olaparib alone, or standard of care chemotherapy in women with recurrent platinum-resistant or -refractory ovarian, fallopian tube, or primary peritoneal cancer (102). Finally, the randomized phase III ICON 9 trial, assesses the efficacy, safety and tolerability of maintenance olaparib in combination with cediranib compared to maintenance olaparib alone following a response to platinum-based chemotherapy in women with relapsed platinum-sensitive ovarian, fallopian tube or peritoneal cancer (103).
In clinical practice, beyond anti-angiogenics, and immune checkpoint inhibitors, we aim to reduce overlapping toxicities by combining PARPis separately with several other agents, such as phosphoinositide 3-kinase (PI3K), protein kinase B (AKT), mammalian target of rapamycin (mTOR), WEE1, mitogen-activated protein kinase kinase (MEK), and cyclin dependent kinase (CDK) 4/6 inhibitors (104).
Recommendations on testing for EOC
Summarizing the available evidence on germline and tumor testing for EOC patients, and having as a goal to target both the hereditary and therapeutic component of ovarian cancer, germline genetic testing should be offered to all women diagnosed with EOC. This referral should be done irrespectively of their age at diagnosis or relevant family history, while the test ordered should be able to detect damaging variants in all genes associated to ovarian cancer susceptibility and not be restricted to BRCA1 and BRCA2 testing. Subsequently, it is recommended to order tumor testing, at least for BRCA1 and BRCA2 genes, in all women tested negative for germline PVs. It would be of the greatest benefit of the patients that the test(s) is offered at the time of initial diagnosis (105).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Stergios Boussios and Nicholas Pavlidis) for the series “Ovarian Cancer: State of the Art and Perspectives of Clinical Research” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1422). The series “Ovarian Cancer: State of the Art and Perspectives of Clinical Research” was commissioned by the editorial office without any funding or sponsorship. Dr. FF reports personal fees from ASTRAZENECA, grants from ASTRAZENECA, during the conduct of the study. Dr. CP reports other from ASTRAZENECA, other from PFIZER, other from Novartis, other from Genesis, other from MSD, other from Amgen, other from Merck, other from Roche, grants from BMS, grants from Roche, during the conduct of the study. Dr. MP has no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lheureux S, Gourley C, Vergote I, et al. Epithelial ovarian cancer. Lancet 2019;393:1240-53. [Crossref] [PubMed]
- Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2019;144:1941-53. [Crossref] [PubMed]
- Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284-96. [Crossref] [PubMed]
- Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2014. Bethesda, MD: National Cancer Institute. Updated April 2, 2018. Available online: https://seer.cancer.gov/archive/csr/1975_2014/
- Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: a review. Cancer Biol Med 2017;14:9-32. [Crossref] [PubMed]
- Gilks CB, Prat J. Ovarian carcinoma pathology and genetics: recent advances. Hum Pathol 2009;40:1213-23. [Crossref] [PubMed]
- Banerjee S, Kaye SB. New strategies in the treatment of ovarian cancer: current clinical perspectives and future potential. Clin Cancer Res 2013;19:961-8. [Crossref] [PubMed]
- Testa U, Petrucci E, Pasquini L, et al. Ovarian cancers: genetic abnormalities, tumor heterogeneity and progression, clonal evolution and cancer stem cells. Medicines (Basel) 2018;5:16. [Crossref] [PubMed]
- Prat J, D'Angelo E, Espinosa I. Ovarian carcinomas: at least five different diseases with distinct histological features and molecular genetics. Hum Pathol 2018;80:11-27. [Crossref] [PubMed]
- Papadimitriou M, Mountzios G, Papadimitriou CA. The role of PARP inhibition in triple-negative breast cancer: Unraveling the wide spectrum of synthetic lethality. Cancer Treat Rev 2018;67:34-44. [Crossref] [PubMed]
- Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 2013;5:a012740. [Crossref] [PubMed]
- Wu X, Ranganathan V, Weisman DS, et al. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 2000;405:477-82. [Crossref] [PubMed]
- Sartori AA, Lukas C, Coates J, et al. Human CtIP promotes DNA end resection. Nature 2007;450:509-14. [Crossref] [PubMed]
- Talens F, Jalving M, Gietema JA, et al. Therapeutic targeting and patient selection for cancers with homologous recombination defects. Expert Opin Drug Discov 2017;12:565-81. [Crossref] [PubMed]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 2011;12:90-103. [Crossref] [PubMed]
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 2006;25:5864-74. [Crossref] [PubMed]
- Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin neocarzinostastin and other enediynes. Mutat Res 1996;355:71-89. [Crossref] [PubMed]
- Chow MS, Liu LV, Solomon EI. Further insights into the mechanism of reaction of activated bleomycin with DNA. Proc Natl Acad Sci U S A 2008;105:13241-5. [Crossref] [PubMed]
- Ma Q, Akiyama Y, Xu Z, et al. Identification and cleavage site analysis of DNA sequences bound strongly by bleomycin. J Am Chem Soc 2009;131:2013-22. [Crossref] [PubMed]
- Champoux JJ. DNA topoisomerases: structure, function and mechanism. Annu Rev Biochem 2001;70:369-413. [Crossref] [PubMed]
- Wang JC. Cellular role of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 2002;3:430-40. [Crossref] [PubMed]
- Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer 2006;6:789-802. [Crossref] [PubMed]
- Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer 2009;9:338-50. [Crossref] [PubMed]
- Rouleau M, Patel A, Hendzel MJ, et al. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 2010;10:293-301. [Crossref] [PubMed]
- Ohmoto A, Yachida S. Current status of poly(ADP-ribose) polymerase inhibitors and future directions. Onco Targets Ther 2017;10:5195-208. [Crossref] [PubMed]
- Hong SJ, Dawson TM, Dawson VL. Nuclear and Mitochondrial Conversations in Cell Death. Trends Pharmacol Sci 2004;25:259-64. [Crossref] [PubMed]
- Yu SW, Wang H, Poitras MF, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent Cell Death by Apoptosis-Inducing Factor. Science. 2002;297:259-63. [Crossref] [PubMed]
- Bridges CB. The origin of variations in sexual and sexlimited characters. Am Nat 1922;56:51-63. [Crossref]
- Nijman SMB. Synthetic lethality: General principles, utility and detection using genetic screens in human cells. FEBS Lett 2011;585:1-6. [Crossref] [PubMed]
- O'Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat Rev Genet 2017;18:613-23. [Crossref] [PubMed]
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913-7. [Crossref] [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [Crossref] [PubMed]
- Macpherson P, Barone F, Maga G, et al. 8-oxoguanine incorporation into DNA repeats in vitro and mismatch recognition by MutSalpha. Nucleic Acids Res 2005;33:5094-105. [Crossref] [PubMed]
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res 2008;18:85-98. [Crossref] [PubMed]
- Pal T, Permuth-Wey J, Sellers TA. A review of the clinical relevance of mismatch-repair deficiency in ovarian cancer. Cancer 2008;113:733-42. [Crossref] [PubMed]
- Xiao X, Melton DW, Gourley C. Mismatch repair deficiency in ovarian cancer-Molecular characteristics and clinical implications. Gynecol Oncol 2014;132:506-12. [Crossref] [PubMed]
- Li SK, Martin A. Mismatch repair and colon cancer: mechanisms and therapies explored. Trends Mol Med 2016;22:274-89. [Crossref] [PubMed]
- Lynch HT, Casey MJ, Snyder CL, et al. Hereditary ovarian carcinoma: heterogeneity, molecular genetics, pathology, and management. Mol Oncol 2009;3:97-137. [Crossref] [PubMed]
- Watson P, Vasen HFA, Mecklin JP, et al. The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. Int J Cancer 2008;123:444-9. [Crossref] [PubMed]
- Zhao C. Prognostic values of DNA mismatch repair genes in ovarian cancer patients treated with platinum-based chemotherapy. Arch Gynecol Obstet 2018;297:153-9. [Crossref] [PubMed]
- Walsh T, Casadei S, Lee MK, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:18032-7. [Crossref] [PubMed]
- Jervis S, Song H, Lee A, et al. Ovarian cancer familial relative risks by tumour subtypes and by known ovarian cancer genetic susceptibility variants. J Med Genet 2014;51:108-13. [Crossref] [PubMed]
- La Vecchia C. Ovarian cancer: epidemiology and risk factors. Eur J Cancer Prev 2017;26:55-62. [Crossref] [PubMed]
- Flaum N, Crosbie EJ, Edmondson RJ, et al. Epithelial ovarian cancer risk: a review of the current genetic landscape. Clin Genet 2020;97:54-63. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609-15. Erratum in: Nature. 2012 Oct 11;490(7419):298. [Crossref] [PubMed]
- Polak P, Kim J, Braunstein LZ, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet 2017;49:1476-86. [Crossref] [PubMed]
- Alsop K, Fereday S, Meldrum C, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian ovarian cancer study group. J Clin Oncol 2012;30:2654-63. [Crossref] [PubMed]
- Lippi G, Mattiuzzi C, Montagnana M. BRCA population screening for predicting breast cancer: for or against? Ann Transl Med 2017;5:275. [Crossref] [PubMed]
- Manickam K, Buchanan AH, Schwartz MLB, et al. Exome sequencing-based screening for BRCA1/2 expected pathogenic variants among adult biobank participants. JAMA Netw Open 2018;1:e182140. [Crossref] [PubMed]
- Konstantinopoulos PA, Matulonis UA. Targeting DNA damage response and repair as a therapeutic strategy for ovarian cancer. Hematol Oncol Clin North Am 2018;32:997-1010. [Crossref] [PubMed]
- Norquist BM, Harrell MI, Brady MF, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol 2016;2:482-90. [Crossref] [PubMed]
- Lu HM, Li S, Black MH, et al. Association of breast and ovarian cancers with predisposition genes identified by large-scale sequencing. JAMA Oncol 2019;5:51-7. [Crossref] [PubMed]
- Zhang S, Royer R, Li S, et al. Frequencies of BRCA1 and BRCA2 mutations among 1,342 unselected patients with invasive ovarian cancer. Gynecol Oncol 2011;121:353-7. [Crossref] [PubMed]
- Song H, Cicek MS, Dicks E, et al. The contribution of deleterious germline mutations in BRCA1, BRCA2 and the mismatch repair genes to ovarian cancer in the population. Hum Mol Genet 2014;23:4703-9. [Crossref] [PubMed]
- Ramus SJ, Harrington PA, Pye C, et al. Contribution of BRCA1 and BRCA2 mutations to inherited ovarian cancer. Hum Mutat 2007;28:1207-15. [Crossref] [PubMed]
- Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 2017;317:2402-16. [Crossref] [PubMed]
- Kotsopoulos J, Gronwald J, Karlan B, et al. Age-specific ovarian cancer risks among women with a BRCA1 or BRCA2 mutation Gynecol Oncol 2018;150:85-91. [Crossref] [PubMed]
- Kauff ND, Satagopan JM, Robson ME, et al. Risk-reducing salpingo-oophorectomy in women with a BRCA1 or BRCA2 mutation. N Engl J Med 2002;346:1609-15. [Crossref] [PubMed]
- Finch AP, Lubinski J, Moller P, et al. Impact of oophorectomy on cancer incidence and mortality in women with a BRCA1 or BRCA2 mutation J Clin Oncol 2014;32:1547-53. [Crossref] [PubMed]
- Domchek SM, Rebbeck T. Prophylactic oophorectomy in women at increased cancer risk. Curr Opin Obstet Gynecol 2007;19:27-30. [Crossref] [PubMed]
- Lilyquist J, La Duca H, Polley E, et al. Frequency of mutations in a large series of clinically ascertained ovarian cancer cases tested on multi-gene panels compared to reference controls. Gynecol Oncol 2017;147:375-80. [Crossref] [PubMed]
- Loveday C, Turnbull C, Ramsay E, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet 2011;43:879-82. [Crossref] [PubMed]
- Konstanta I, Fostira F, Apostolou P, et al. Contribution of RAD51D germline mutations in breast and ovarian cancer in Greece. J Hum Genet 2018;63:1149-58. [Crossref] [PubMed]
- NCCN Guidelines for Genetic/Familial High-Risk Assessment: Breast and Ovarian. Version 3.2019. Available online: https://www2.trikobe.org/nccn/guideline/gynecological/english/genetic_familial.pdf
- Manchanda R, Legood R, Antoniou AC, et al. Specifying the Ovarian Cancer Risk Threshold of 'Premenopausal Risk-Reducing Salpingo-Oophorectomy' for Ovarian Cancer Prevention: A Cost-Effectiveness Analysis. J Med Genet 2016;53:591-9. [Crossref] [PubMed]
- Weber-Lassalle N, Hauke J, Ramser J, et al. BRIP1 loss-of-function mutations confer high risk for familial ovarian cancer, but not familial breast cancer. Breast Cancer Res 2018;20:7. [Crossref] [PubMed]
- Ramus SJ, Song H, Dicks E, et al. Germline Mutations in the BRIP1, BARD1, PALB2, and NBN Genes in Women With Ovarian Cancer. J Natl Cancer Inst 2015;107:djv214. [Crossref] [PubMed]
- Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med 2014;371:497-506. [Crossref] [PubMed]
- Yang X, Leslie G, Doroszuk A, et al. Cancer risks associated with germline PALB2 pathogenic variants: an international study of 524 families. J Clin Oncol 2020;38:674-85. [Crossref] [PubMed]
- Isaac D, Karapetyan L, Tamkus D. Association of germline PALB2 mutation and response to platinum-based chemotherapy in metastatic breast cancer: a case series. JCO Precis Oncol 2018;2:1-5. [Crossref]
- Mills AM, Longacre TA. Lynch syndrome screening in the gynecologic tract: current state of the art. Am J Surg Pathol 2016;40:e35-44. [Crossref] [PubMed]
- Bonadona V, Bonaïti B, Olschwang S, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA 2011;305:2304-10. [Crossref] [PubMed]
- Dowty JG, Win AK, Buchanan DD, et al. Cancer risks for MLH1 and MSH2 mutation carriers. Hum Mutat 2013;34:490-7. [Crossref] [PubMed]
- Møller P, Seppala T, Bernstein I, et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut 2017;66:464-72. [Crossref] [PubMed]
- Dominguez-Valentin M, Sampson JR, Seppälä TT, et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database. Genet Med 2020;22:15-25. [Crossref] [PubMed]
- Ryan NAJ, Evans DG, Green K, et al. Pathological features and clinical behavior of Lynch syndrome-associated ovarian cancer. Gynecol Oncol 2017;144:491-5. [Crossref] [PubMed]
- Helder-Woolderink JM, Blok EA, Vasen HFA, et al. Ovarian cancer in Lynch syndrome; a systematic review. Eur J Cancer 2016;55:65-73. [Crossref] [PubMed]
- Hennessy BTJ, Timms KM, Carey MS, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol 2010;28:3570-6. [Crossref] [PubMed]
- Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 2014;20:764-75. [Crossref] [PubMed]
- Jacobs S, Thompson ER, Nannya Y, et al. Genome‐wide, high‐resolution detection of copy number, loss of heterozygosity, and genotypes from formalin‐fixed, paraffin‐embedded tumor tissue using microarrays. Cancer Res 2007;67:2544-51. [Crossref] [PubMed]
- Bouwman P, Jonkers J. Molecular pathways: how can BRCA-mutated tumors become resistant to PARP inhibitors? Clin Cancer Res 2014;20:540-7. [Crossref] [PubMed]
- Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med 2016;375:2154-64. [Crossref] [PubMed]
- Swisher EM, Lin KK, Oza AM, et al. Rucaparib in platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international multicentre, open-label, phase 2 trial. Lancet Oncol 2017;18:75-87. [Crossref] [PubMed]
- Moore K, Colombo N, Scambia G, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2018;379:2495-505. [Crossref] [PubMed]
- Pujade-Lauraine E, Ledermann JA, Selle F, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 2017;18:1274-84. [Crossref] [PubMed]
- Penson RT, Villalobos Valencia R, Cibula D, et al. Olaparib monotherapy versus (vs) chemotherapy for germline BRCA-mutated (gBRCAm) platinum-sensitive relapsed ovarian cancer (PSR OC) patients (pts): Phase III SOLO3 trial. J Clin Oncol 2019;37:abstr 5506.
- González-Martín A, Pothuri B, Vergote I, et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med 2019;381:2391-402. [Crossref] [PubMed]
- Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:1949-61. [Crossref] [PubMed]
- Ray-Coquard I, Pautier P, Pignata S, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 2019;381:2416-28. [Crossref] [PubMed]
- Ledermann JA, Harter P, Gourley C, et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol 2016;17:1579-89. [Crossref] [PubMed]
- Ledermann JA, Pujade-Lauraine E. Olaparib as maintenance treatment for patients with platinum-sensitive relapsed ovarian cancer. Ther Adv Med Oncol 2019;11:1758835919849753. [Crossref] [PubMed]
- Matsumoto K, Nishimura M, Onoe T, et al. PARP inhibitors for BRCA wild type ovarian cancer; gene alterations, homologous recombination deficiency and combination therapy. Jpn J Clin Oncol 2019;49:703-7. [Crossref] [PubMed]
- Huang J, Wang L, Cong Z, et al. The PARP1 inhibitor BMN 673 exhibits immunoregulatory effects in a Brca1(-/-) murine model of ovarian cancer. Biochem Biophys Res Commun 2015;463:551-6. [Crossref] [PubMed]
- Drew Y, Kaufman B, Banerjee S, et al. Phase II study of olaparib + durvalumab (MEDIOLA): Updated results in germline BRCA-mutated platinum-sensitive relapsed (PSR) ovarian cancer (OC). Ann Oncol 2019;30:v485-6. [Crossref]
- Konstantinopoulos PA, Waggoner S, Vidal GA, et al. Single-arm phases 1 and 2 trial of niraparibin combination with pembrolizumab in patients with recurrent platinum-resistant ovarian carcinoma. JAMA Oncol 2019;5:1141-9. [Crossref] [PubMed]
- Hardy-Bessard A-C, Moore KN, Mirza MR, et al. ENGOT-OV44/FIRST study: a randomized, double-blind, adaptive, phase 3 study of standard of care (SOC) platinum-based therapy ± dostarlimab followed by niraparib ± dostarlimab maintenance as first-line (1L) treatment of stage 3 or 4 ovarian cancer (OC). J Clin Oncol 2020;38:TPS6101. [Crossref]
- A Study in Ovarian Cancer Patients Evaluating Rucaparib and Nivolumab as Maintenance Treatment Following Response to Front-Line Platinum-Based Chemotherapy (ATHENA). Available online: https://clinicaltrials.gov/ct2/show/NCT03522246
- Chan N, Bristow RG. ‘Contextual’ synthetic lethality and/or loss of heterozygosity: tumor hypoxia and modification of DNA repair. Clin Cancer Res 2010;16:4553-60. [Crossref] [PubMed]
- Mirza MR, Åvall Lundqvist E, Birrer MJ, et al. Niraparib Plus Bevacizumab Versus Niraparib Alone for Platinum-Sensitive Recurrent Ovarian Cancer (NSGO-AVANOVA2/ENGOT-ov24): A Randomised, Phase 2, Superiority Trial. Lancet Oncol 2019;20:1409-19. [Crossref] [PubMed]
- Lee J-M, Moore RG, Ghamande SA, et al. Cediranib in combination with olaparib in patients without a germline BRCA1/2 mutation with recurrent platinum-resistant ovarian cancer: Phase IIb CONCERTO trial. J Clin Oncol 2020;38:abstr 6056.
- Nicum S, Strauss VY, McGregor N, et al. OCTOVA: A randomised phase II trial of olaparib, chemotherapy, or olaparib and cediranib in patients with BRCA-mutated platinum-resistant ovarian cancer. Ann Oncol 2017;28:v330-54. [Crossref]
- Cediranib Maleate and Olaparib or Standard Chemotherapy in Treating Patients With Recurrent Platinum-Resistant or -Refractory Ovarian, Fallopian Tube, or Primary Peritoneal Cancer. Available online: https://www.clinicaltrials.gov/ct2/show/NCT02502266
- Study Evaluating the Efficacy of Maintenance Olaparib and Cediranib or Olaparib Alone in Ovarian Cancer Patients (ICON9). Available online: https://clinicaltrials.gov/ct2/show/NCT03278717
- Boussios S, Karihtala P, Moschetta M, et al. Combined Strategies with Poly (ADP-Ribose) Polymerase (PARP) Inhibitors for the Treatment of Ovarian Cancer: A Literature Review. Diagnostics (Basel) 2019;9:87. [Crossref] [PubMed]
- Konstantinopoulos PA, Norquist B, Lacchetti C, et al. Germline and Somatic Tumor Testing in Epithelial Ovarian Cancer: ASCO Guideline. J Clin Oncol 2020;38:1222-45. [Crossref] [PubMed]


