DeepLN: an artificial intelligence-based automated system for lung cancer screening
Introduction
Lung cancer is responsible for more deaths worldwide than any other cancer (1). In 2018 alone, 2.1 million people were newly diagnosed with lung cancer, and 1.8 million people died from the disease. In the next few decades, it is expected that the incidence of lung cancer in China will rise, owing to the high rate of smoking in the population (1). At only 18%, the 5-year survival rate of lung cancer is low (2), which can mainly be explained by most patients already having advanced-stage disease at the time they are first diagnosed. Fortunately, by expanding standard screening programs, lung cancer may be diagnosed at an earlier stage, which has been evidenced by decreases in disease mortality in both the United States and Japan (2). Low-dose computed tomography (LDCT) of the chest is considered to be the primary method for lung cancer screening, especially for patients who have an elevated risk of lung cancer (2,3). By the effective evaluation and follow-up of lung nodules (LNs) found on LDCT images during screening (2,3), the persons can obtain timely diagnostic suggestions. Thus, the accurate detection of LNs and identification of the probability of malignancy based on LDCT images hold significant value in the early diagnosis of lung cancer and may improve the survival of lung cancer patients.
In the last two decades, many computer-aided detection (CAD) techniques for the automated detection of LNs have been presented and shown to improve the work efficiency and performance of radiologists (4). Generally, these methods were based on hand-crafted features and classifiers derived from traditional machine learning techniques. However, hand-crafted features still rely on human interpretation of medical images and so their accuracy varies widely. Recently, deep neural networks (DNNs), a popular artificial intelligence (AI) algorithm, have equaled or outperformed human experts in several medical image analysis tasks, such as the detection of diabetic retinopathy (5,6), breast cancer diagnosis (7,8), electrocardiogram (ECG) data analysis (9), and other tasks (10,11). In 2016, the LUNA16 challenge (12), which called for algorithms to detect LNs in a dataset of 888 computed tomography (CT) images with annotation from four radiologists, the method with the best performance applied a convolutional neural network (CNN) model. Inspired by the promising performance of CNNs, an increasing number of DNNs have been applied to detect LNs in CT images, including a region-based CNN (R-CNN) (13), a spatial pyramid pooling (SPP) network (14), a fast R-CNN (15), and a faster R-CNN (a region proposal network, RPN) (16). Setio et al. (17) presented a CAD system for LN detection by using a multi-view CNN. Dou et al. developed a three-dimensional (3D) CNN (ConvNets) (18) to automatically detect LNs on CT scans by first training a fully convolutional network (FCN) to screen LN candidates, then designing a hybrid-loss residual network to lower the false positive rate. In a later report, they presented a 3D CNN framework formed through the integration of multilevel contextual information to further reduce the rate of false positives and to accurately discriminate true LNs (19). Ding et al. (20) adopted the faster R-CNN (16) for LN candidate discrimination and reduced the rate of false positives using a 3D CNN. These presented models (17-20) were evaluated on the LUNA16 dataset (12,21) and showed good performances. Xu et al. (22) proposed a novel non-maximum suppression-based ensemble strategy to synthesize the results from multiple neural network models on a multiresolution LDCT image dataset. The LN detection result was considered acceptable.
Another key component of lung cancer screening is evaluating the risk of malignant LNs. The challenge of distinguishing malignant LNs from benign LNs has drawn attention from many researchers. Han et al. (23) used a support vector machine classifier to differentiate between malignant LNs and benign LNs based on three texture-related features. Shen et al. (24) proposed a multi-crop CNN model to classify the risk of LN malignancy as high- or low-suspicion nodules. Liu et al. (25) proposed a multi-task deep learning model for score grading and LN classification. Xie et al. proposed a series of methods (26-29) for identifying benign and malignant LNs on chest CT images and validated these methods on the Lung Image Database Consortium image collection (LIDC-IDRI) (21). In their latest work (29), to address the issue of limited training data, a semi-supervised adversarial classification model was proposed for classifying LN malignancy. To predict the malignancy of LNs, Liao et al. adopted a 3D deep leaky noisy-or network, and the developed model exhibited a high area under the receiver operator curve (AUC) (30).
The majority of these earlier studies focused on either detecting or classifying LNs. According to the latest review of pulmonary medical imaging (31), which covers most of the methods of LN detection, classification, and segmentation and the available public datasets, only one study has resulted in a comprehensive lung cancer screening system through which clinical decisions can be made based on LDCT images directly. In this study (32), Ardila et al. proposed an end-to-end method with two CNN models that detected the cancer candidate region in a CT volume and then output the case-level lung cancer malignancy scores. However, their system did not detect LNs but instead focused on detecting suspicious regions of interest (i.e., the cancer candidate regions) to assign a case-level malignancy score. However, it is necessary to detect all suspicious LNs, because the clinical course of treatment is determined based on the findings from LDCT images captured at the initial and follow-up screenings. Furthermore, the characteristics of LN, such as the size (including the largest LN diameter and volume), density, the involved lung lobe (33,34) are also important radiologic factors for clinical decision-making. Thus, a comprehensive lung cancer identification system is needed for lung cancer screening with LDCT.
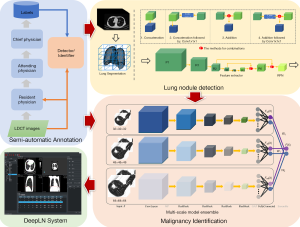
In this paper, we propose a complete framework based on the DNNs (as shown in Figure 1), including an accurate and convenient annotation tool, two DNN-based models for LN detection, and malignancy classification, and an automated lung cancer screening system. First, a semi-automatic annotation strategy was presented to collect LDCT images and generate training labels of LNs and the degree of malignancy based on the confirmed pathological reports. Then, the DNN-based LN detector and malignancy classifier were proposed and trained on the self-constructed dataset to detect all suspicious LNs from chest LDCT images and identify them as benign or malignant LNs. The core properties of LNs, including the largest diameter and volume, were also calculated to supply more information to radiologists. The proposed models were integrated into a system referred to as DeepLN, which was developed by using Qt and the Microsoft Visual Studio platform. The DeepLN system can be easily set up on a computer and connected to a picture archiving and communication system (PACS) to automatically obtain LDCT images and rapidly return the diagnostic results. This developed system was retrospectively evaluated on several datasets to validate its performance.
Methods
As shown in Figure 1, the proposed framework comprises three main modules: the semi-automatic annotation module, the automated LN detection module, and the malignancy classification module. The first module was designed to collect LDCT images and to obtain the training labels for the other two modules. Next, as shown in Figure 1, two DNN models were designed: one for the LN detection module to accurately detect all of the suspicious LNs located in the lung region and output the 3D positions within the volumetric LDCT images and the LN size, and another for the malignancy identification module to classify the LNs as benign or malignant LN to yield a diagnostic result.
The second and third modules were integrated into the DeepLN system to output diagnostic results from the images automatically and accurately. The following subsections describe the three modules and the developed system in detail.
Image labelling
While deep learning algorithms have achieved much success in many medical applications, a large number of labeled images were needed as training data, which may not be available for most medical applications. In AI-based studies, medical image annotation is one of the most crucially important steps. A web-based LN-annotation tool named DeepLNAnno was proposed in our earlier study (35) to annotate LN regions to ease an automated LN detection. The tool is easy to use, when the annotators find a LN on the CT images, they only need use the mouse to draw a rectangle around it, the location and size of the corresponding rectangle will be recorded as the label for LN detection task. In addition, in this study, this tool was extended through the addition of a function to annotate benign and malignant LNs, in this way the labels for LN identification task can be obtained. The CT images in this study were collected from patients who admitted to the collaborated hospital or are at follow-up. For the patients who have been treated, the screening images before treatment were included. In addition, to confirm the accuracy of the benign/malignancy annotation for each LN, the pathology reports obtained from the pathology department were used as the annotating reference.
The annotation was carried out in collaboration with a medical group including PhD. candidates, resident physicians, attending physicians, and chief physicians from the departments of respiratory, radiology, and thoracic surgery from West China Hospital of Sichuan University (a Grade-III hospital in China). A three-tier annotation procedure was constructed: the first round of annotations was provided by the PhD. candidates and resident physicians; next, the attending physicians (each of whom had at least five years of clinical experience) reviewed the results to correct the incorrectly identified LNs and supplement the LNs that had been missed; and finally, the final review was performed by chief physicians with at least five years of medical experience. This procedure ensured the accuracy of the labels, although its labor-intensive nature and the significant human effort it demands could prove expensive.
Following the human annotation of images from 500 patients, a semi-automated procedure was also designed, by training a 3D CNN model on this manually annotated data. The model was then applied to the newly collected data to identify reference LNs in the first round of annotation. Through this, the semi-automated function would improve the annotation efficiency by aiding the subsequent annotation work and reducing the rate of missed LNs in the first round of annotation.
LN detection
In the LN detection module, the lung region was initially segmented into a series of binary images through the application of a traditional thresholding method. These images were then used as masks to segment lung regions from corresponding volumetric LDCT images as the input, I, to the detection model. The output was a four-element vector, [x, y, z, d], in which x, y, and z represent the coordinates of the center of an LN in the 3D volumetric LDCT images and d represents the largest diameter of the LN. The 3D detection network model was constructed based on the RPN (16) and residual networks (36), as shown in Figure 1 (top right). The network backbone starts with 2 convolutional layers with a 3×3×3 kernel size (i.e., P1 in Figure 1), followed by 6 residual blocks, Ri(i = 1…6) and an RPN. The max-pooling (MP) operation was applied to halve the spatial size of the feature map before inputting into the four residual blocks. The first four residual blocks contained two convolutional layers, followed by a batch-normalization layer and a ReLU activation layer. Then, the spatial size of the feature maps was doubled by applying a deconvolution operation before the final two residual blocks. Furthermore, feature maps with the same spatial size were combined for LN detection (i.e., the feature maps R2 and R6 and those of R3 and R5). As shown in Figure 1, four combination strategies were designed to improve the detection results: concatenation, concatenation with Conv1×1×1, element-wise addition, and element-wise addition with Conv1×1×1. The feature maps were also doubled in size before combination to make sure that the number of feature maps could be maintained by element-wise addition. After the combination step, the LNs were detected by inputting the feature map into the RPN. The end-to-end detector comprised a total of 22 layers, including convolutional layers, pooling layers, and a region proposal layer. The output of the RPN was a five-element vector [pi,xi, yi, zi, di], in which pi denotes the probability of a candidate LN, (xi, yi, zi) are the center coordinates and di is the diameter.
Malignancy identification
The detected LNs were then transferred to the identification module to obtain the malignancy risk diagnosis. The architecture of the neural network model for malignancy identification is shown in Figure 1 (bottom right). To improve the training efficiency and confirm the experimental accuracy, three identification models with the same network structure and different input sizes were constructed. The LNs detected were cropped from raw LDCT images, and 3D volumes of 32×32×32, 48×48×48, and 64×64×64 voxels around or near the LN center were preprocessed and saved to train three identification models, respectively. The results obtained from the three models were later combined to obtain a final ensemble result for malignancy evaluation. The construction of the identification model started with a convolutional layer and a MP layer. Next, the extracted features were passed through four residual blocks, with a second MP layer added after the first three residual blocks. The features extracted from the last residual block were flattened into a feature vector by a global average pooling layer. Finally, a fully connected layer and an output layer completed the model. Thus, this was a light model with a total of 10 layers. Data augmentation operations of flipping and translation were employed to increase the sample diversity to train the model effectively on the limited dataset. The weighted cross-entropy loss (CEL) function was applied to counter the problem posed by imbalanced category distribution.
The DeepLN system
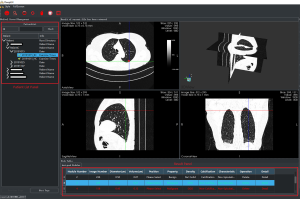
As shown in Figure 2, an interactive system with a visual graphical user interface (GUI) was constructed to aid the radiologists in lung cancer screening. The two proposed DNN-based models were then integrated into the system, which included two panels and four viewers to display the LDCT images (an axial slice viewer, a sagittal slice viewer, a coronal slice viewer, and a 3D viewer). The system was integrated with the PACS in West China Hospital to automatically obtain the LDCT data of the patients who were examined. The former LDCT images could also be downloaded automatically when the system was first installed. In this way, all the data could be visualized by the doctor for further analysis during follow-up visits. As shown in Figure 3, an interface was designed to visualize the LDCT images collected at various times. The obtained data was displayed in the patient list panel (Figure 2). The obtained LDCT images were sequentially input into the detection and identification models, and the results were shown in the result panel, including the LN index, slice index of the image in the LDCT dataset (on which LN has the largest diameter), and the estimated volume, diameter, involved lung lobe, malignancy risk, density, and other characteristics of the detected LN. These details could offer significant value in aiding the radiologist in making a follow-up program or a treatment plan.
Results
Datasets
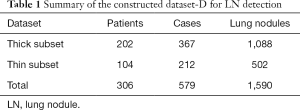
The data used in this study were collected from West China Hospital, Sichuan University, China. All CT screening images were collected from patients who were either diagnosed with lung cancer or during follow-up of LNs. As a result, some patients had more than one set of LDCT images available and included in the dataset. For patients who had been diagnosed with lung cancer, only the LDCT images acquired before the treatment were included. Furthermore, guidelines were provided during the annotation to obtain a high-quality dataset and ensure uniformity among the radiologists. In essence, all nodules observed inside the lung region were labeled regardless of size (i.e., even those with diameters less than 5 mm, for which annual LDCT screening is indicated (2,3) to ensure follow-up for benign LNs such as inflammatory or calcified LNs. LNs in the pleura were disregarded. Cases in which too many LNs were present to allow for accurate annotation were removed from the study. Finally, a dataset consisting of 1,590 LNs in 579 case sets of images from 306 patients was constructed and annotated, as summarized in Table 1. Denoted as dataset-D, this dataset included images captured with different thick sections. In general, if a patient has a high risk of lung cancer, thin-section scans are required, while thick-section LDCT scans with lower radiation are used for initial screening as the cancer risk associated with radiation cannot be neglected (37). As shown in Table 1, CT images with 1.5 mm- and 5 mm-thick sections were collected and denoted as the thin subset and thick subset, respectively.
Full table
The labeled LNs were further annotated as either benign or malignant. Although several diagnostic tests are commonly used to determine malignancy risk in CT-screen-detected LNs in clinical practice (33), surgical excisional biopsies are the gold-standard for establishing a definitive diagnosis. Thus, in this study, the annotation of malignancy classification was verified based on the pathology results from surgical biopsies. In this study, pathologic results were available for 549 of the study cases. As shown in Table 2, a total of 766 LNs were annotated, including 567 benign LNs and 199 malignant LNs; this dataset was denoted as dataset-I.

Full table
Lung cancer detection performance
The training and testing of both models for lung cancer identification were conducted on a workstation with an Ubuntu server 14.04 system and four 24 GB NVIDIA Titan RTX cards. The designed models were implemented using PyTorch-v1.0.1 and Python37.
The LN detection model was trained by using stochastic gradient descent (SGD) with a first learning rate of 0.01 and a momentum of 0.9. 3D volumetric patches were cropped from the CT images and input into the model; the input size was empirically (22) set as 128×128×128, and concatenation was employed as a combination method. In this study, two subsets with different section thicknesses were included in the dataset. Instead of simple resampling, each detection model was trained on each subset separately to obtain better performance. Dataset-D was split into two subsets for the detection task: the training set comprised 80% of the cases, and the testing set comprised the remaining 20%. The training set included 294 cases from the thick subset and 169 cases from the thin subset, while the testing set included 73 cases from the thick subset and 43 cases from the thin subset. The model performance in the detection task was evaluated using two commonly used metrics: the sensitivity, which represents the true positive rate (TPR), and the free-response receiver operating characteristic (FROC), which is a measure of the false-positive (FP) reduction and can further validate the sensitivity of the model. The TPR was defined as TP/(TP + FN), in which TP is the number of true positives, and FN is the number of false negatives. The average FROC value at 1/8, 1/4, 1/2, 1, 2, 4, 8 false positive per scan is calculated.
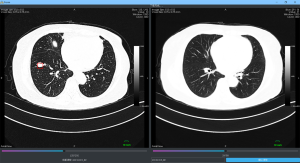
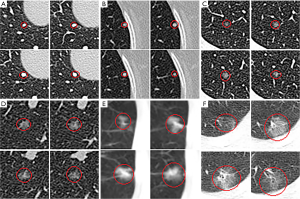
The experimental results are shown in Table 3. The sensitivity of the thin-subset was 96.5% and that of the thick-subset was 89.6%, which is comparable to those in earlier studies (22). Also, the detected nodules are shown in Figure 4. It is demonstrated that the different types of nodules, such as solid LN, partially solid LN, and ground-glass nodule, have important clinical values and can be effectively detected.
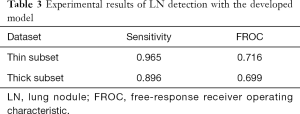
Full table
LN malignancy classification performance
The performance of the model in classifying benign and malignant LNs was evaluated, and the results are shown in Table 4. This study employed the commonly used evaluation metrics, including accuracy, sensitivity/recall, specificity, precision with a cut-off value of 0.5, AUC, and F1-score. The model was trained for 120 epochs, and its parameters were saved at the end of each epoch. The batch size during the training was 32. The experimental results of the 10-fold cross-validation are shown in Table 4. The experimental results demonstrate that the model performance was influenced by the size of the 3D volumetric LN cropped from raw CT data, and it is difficult to theoretically determine the appropriate input size. The results showed that each metric was optimized with either the smallest or largest input size. These findings imply that the results can be substantially improved by the ensemble strategy. Hence, the ensemble strategy proposed in (38) was employed to combine the results obtained from the three models based on a grid search. As expected, the accuracy in the LN malignancy classification reached 92.46%, which is considered to represent a satisfactory result in medicine.
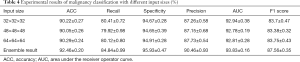
Full table
A retrospective comparison with clinical experts
To objectively validate the performance of the proposed system further, three retrospective comparisons were performed by human clinical experts at West China Hospital. The study cases and corresponding medical image diagnostic reports were collected, and case-level comparisons with the results from the system were conducted. The results were compared to verify that all of the LNs included in the case report were detected by the proposed system (i.e., there were no false negatives). Even if only one LN was missed by the model, the entire case was designated as a false positive; in other words, the evaluation accuracy was obtained at 1 FP/scan. It should also be specified that only significant and representative LNs in the CT images were presented in the report. This study did not perform comparisons of whether all of the LNs were identified by the system as well as in the case report.
As shown in Table 5, of the 402 study cases included in the first comparison, LNs were missed by the detection model in 9 cases; thus, the accuracy was 97.76%. Then, the detection model was further refined with the LNs included in the medical image diagnostic reports of these 402 cases. In the second comparison, of the 883 cases used, LNs were missed in 8 cases, which is equivalent to an accuracy of 98.98%, exceeding that of the first comparison. With further model refinement based on these data, 99.52% accuracy was obtained in the third comparison with 1,046 cases. Upon review by radiologists, it was noted that the missed LNs were from cases in which the patients were diagnosed with lung squamous carcinoma, which are not clinically considered as nodules, or had atelectasis. In total, 2,336 studies were used for comparison, and the overall accuracy was 99.02%. These results are satisfactory, and the constructed DeepLN system is gaining acceptance from the doctors in West China Hospital.

Full table
Conclusions
As a promising method in AI, deep learning has proven success in many fields of medicine. Lung cancer is responsible for more deaths than any other cancer worldwide, and has a low survival rate. Regular LDCT screening can reduce lung cancer mortality by facilitating the early detection and diagnosis of LNs. In this study, a complete framework for successful lung cancer screening was developed through multidisciplinary collaboration between pulmonologists, pathologists, radiologists, and thoracic surgeons. An effective and easy-to-use system based on DNNs was constructed to evaluate LDCT images to detect LNs and classify them as benign or malignant. This system is expected to better inform the plan for follow-up screening or treatment.
Firstly, an effective annotation strategy was developed because deep learning algorithms require a large amount of labeled data for training. Patient LDCT images were annotated in batches with the aid of a semi-automated annotation tool designed to reduce the annotation workload. The annotation was conducted by a multidisciplinary group of physicians in a three-tiered approach to ensure the correctness of the annotation. Then, a deep convolutional neural network model for LN detection was implemented to extract and combine the high- and low-level features to achieve more exact detection. Then, three light CNN models were proposed and combined in an ensemble manner to evaluate the malignancy of each LN accurately. In this study, the two 3D CNNs were employed as a backbone to extract the LN features from LDCT images and preserve the LN spatial heterogeneity to obtain excellent performance in LN detection and malignancy evaluation. Finally, an interactive system, named DeepLN, with a user-friendly visual GUI interface was developed through the integration of the two DNN models. The system can automatically retrieve patients’ LDCT images from a PACS and rapidly return the diagnostic results.
Furthermore, several characteristics of detected LNs are also displayed intuitively to the user, which can be considered alongside the diagnostic results to further inform the plan for follow-up or treatment. Finally, the developed system was retrospectively evaluated using a large-scale dataset collected from West China Hospital. The accuracy of the system was extremely satisfactory compared to that achieved by human experts.
In future studies, we plan to extend the LN classification task of benign versus malignant to multi-class classification by classifying the benign nodules to hamartomas, bronchial adenomas, papillomas, fibromas and etc. and classifying the malignant nodules to small-cell carcinoma, non-small cell carcinoma, squamous cell carcinoma, adenocarcinoma and etc. The aim is to provide more detailed information with significant clinical value to assist lung cancer screening. Secondly, a more comprehensive analysis of LN characteristics that bear close correlation with the risk of malignancy, such as calcification, lobulation, and speculation, will also be considered. Finally, we plan to collect more training data with accurate labels and to further study the DNN, with the aim of improving the robustness of the model, and multicenter clinical testing will be considered to validate the DeepLN system further.
Acknowledgments
We would like to thank all of the radiologists who dedicated their time and effort to medical image diagnostic reports for the retrospective comparisons.
Funding: This work was supported by the National Key Research and Development Program of China (No. 2018AAA0100201), the Science and Technology Project of Chengdu for Precision Medicine Research (No. 2017-CY02-00030-GX) and by the Major Research plan of the National Natural Science Foundation of China (Grant No. 91859203).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4461). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Wood DE, Kazerooni EA, Baum SL, et al. Lung Cancer Screening, Version 3.2018, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2018;16:412-41. [Crossref] [PubMed]
- Cheng YI, Davies MPA, Liu D, et al. Implementation planning for lung cancer screening in China. Precision Clinical Medicine 2019;2:13-44. [Crossref]
- Bogoni L, Ko JP, Alpert J, et al. Impact of a computer-aided detection (CAD) system integrated into a picture archiving and communication system (PACS) on reader sensitivity and efficiency for the detection of lung nodules in thoracic CT exams. J Digit Imaging 2012;25:771-81. [Crossref] [PubMed]
- Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016;316:2402-10. [Crossref] [PubMed]
- Kermany DS, Goldbaum M, Cai W, et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018;172:1122-31.e9. [Crossref] [PubMed]
- Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women with Breast Cancer. JAMA 2017;318:2199-210. [Crossref] [PubMed]
- McKinney SM, Sieniek M, Godbole V, et al. International evaluation of an AI system for breast cancer screening. Nature 2020;577:89-94. [Crossref] [PubMed]
- Hannun AY, Rajpurkar P, Haghpanahi M, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med 2019;25:65-9. [Crossref] [PubMed]
- Litjens G, Kooi T, Bejnordi B E, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng 2017;19:221-48. [Crossref] [PubMed]
- Setio AA, Traverso A, De Bel T, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med Image Anal 2017;42:1-13. [Crossref] [PubMed]
- Girshick R, Donahue J, Darrell T, et al. Rich feature hierarchies for accurate object detection and semantic segmentation. Computer Vision and Pattern Recognition 2014;1:580-7. [Crossref]
- He K, Zhang X, Ren S, et al. Spatial Pyramid Pooling in Deep Convolutional Networks for Visual Recognition. IEEE Trans Pattern Anal Mach Intell 2015;37:1904-16. [Crossref] [PubMed]
- Girshick R. Fast R-CNN. Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV) 2015:1440-8.
- Ren S, He K, Girshick R, et al. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans Pattern Anal Mach Intell 2017;39:1137-49. [Crossref] [PubMed]
- Setio AA, Ciompi F, Litjens G, et al. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans Med Imaging 2016;35:1160-9. [Crossref] [PubMed]
- Dou Q, Chen H, Yu L, et al. Automated Pulmonary Nodule Detection via 3D ConvNets with Online Sample Filtering and Hybrid-Loss Residual Learning. Med Image Comput Comput Assist Interv 2017:630-8.
- Dou Q, Chen H, Yu L, et al. Multi-level Contextual 3D CNNs for False Positive Reduction in Pulmonary Nodule Detection. IEEE T Bio-Med Eng 2017;64:1558-67.
- Ding J, Li A, Hu Z, et al. Accurate Pulmonary Nodule Detection in Computed Tomography Images Using Deep Convolutional Neural Networks. Med Image Comput Comput Assist Interv 2017:559-67.
- Armato SG 3rd, McLennan G, Bidaut L, et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 2011;38:915-31. [Crossref] [PubMed]
- Xu X, Wang C, Guo J, et al. DeepLN: A framework for automatic lung nodule detection using multi-resolution CT screening images. Knowl-Based Syst 2020. doi: 10.1016/j.knosys.2019.105128. [Crossref]
- Han F, Wang H, Zhang G, et al. Texture Feature Analysis for Computer-Aided Diagnosis on Pulmonary Nodules. J Digit Imaging 2015;28:99-115. [Crossref] [PubMed]
- Shen W, Zhou M, Yang F, et al. Multi-crop Convolutional Neural Networks for lung nodule malignancy suspiciousness classification. Pattern Recognit 2017;61:663-73. [Crossref]
- Liu L, Dou Q, Chen H, et al. MTMR-Net: Multi-task Deep Learning with Margin Ranking Loss for Lung Nodule Analysis. In: Stoyanov D, Taylor Z, Carneiro G, et al., editors. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. DLMIA 2018, ML-CDS 2018, LNCS 2018;11045:74-82.
- Xie Y, Xia Y, Zhang J, et al. Transferable Multi-model Ensemble for Benign-Malignant Lung Nodule Classification on Chest CT. Med Image Comput Comput Assist Interv 2017;10435:656-64.
- Xie Y, Xia Y, Zhang J, et al. Fusing texture, shape and deep model-learned information at decision level for automated classification of lung nodules on chest CT. Inform Fusion 2018;42:102-10. [Crossref]
- Xie Y, Xia Y, Zhang J, et al. Knowledge-based Collaborative Deep Learning for Benign-Malignant Lung Nodule Classification on Chest CT. IEEE Trans Med Imaging 2019;38:991-1004. [Crossref] [PubMed]
- Xie Y, Zhang J, Xia Y. Semi-supervised adversarial model for benign-malignant lung nodule classification on chest CT. Med Image Anal 2019;57:237-48. [Crossref] [PubMed]
- Liao F, Liang M, Li Z, et al. Evaluate the malignancy of pulmonary nodules using the 3D deep leaky noisy-or network. IEEE Trans Neural Netw Learn Syst 2019;30:3484-95. [Crossref] [PubMed]
- Ma J, Song Y, Tian X, et al. Survey on deep learning for pulmonary medical imaging. Front Med 2020;14:450-69. [Crossref] [PubMed]
- Ardila D, Kiraly AP, Bharadwaj S, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 2019;25:954-61. [Crossref] [PubMed]
- Al-Ayoubi AM, Flores RM. Management of CT screen-detected lung nodule: The thoracic surgeon perspective. Ann Transl Med 2016;4:156. [Crossref] [PubMed]
- Pedersen JH, Ashraf H. Implementation and organization of lung cancer screening. Ann Transl Med 2016;4:152. [Crossref] [PubMed]
- Chen S, Guo J, Wang C, et al. DeepLNAnno: a Web-Based Lung Nodules Annotating System for CT Images. J Med Syst 2019;43:197. [Crossref] [PubMed]
- He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. Computer Vision and Pattern Recognition 2016;770-8.
- Mascalchi M, Sali L. Lung cancer screening with low dose CT and radiation harm-From prediction models to cancer incidence data. Ann Transl Med 2017;5:360. [Crossref] [PubMed]
- Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-Learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825-30.
(English Language Editor: J. Chapnick)