
MiR-3121-3p promotes tumor invasion and metastasis by suppressing Rap1GAP in papillary thyroid cancer in vitro
Introduction
Thyroid cancer is the fifth most prevalent cancer type for the estimated new cancer cases in females, accounting for 5% of all new cases in females in the USA in 2017 (1). Among its subtypes, papillary thyroid cancer (PTC) is the most common pathology, accounting for 80% to 85% of all thyroid cancers (2). PTC has relatively better outcome compared with other malignancies. Active observation could be a choice for papillary microcarcinomas. For most papillary cancers that need treatment, surgery is the essential therapy. Further treatments are based on accurate staging and the extent of surgery. Previous studies reported increased recurrence rates with higher clinicopathological risks (3,4). BRAFV600E is the most common and important mutation in papillary thyroid cancer (5). Researches have not only elucidated its role in distinguishing malignant tumors from benign masses but also clarified a strong association between BRAFV600E and poor clinicopathological outcomes of PTC including aggressive pathological features, increased recurrence, loss of radioiodine avidity and treatment failures (6-8). Telomerase reverse transcriptase (TERT) promoter mutation is related to aggressive thyroid tumor characteristics (9). Other than mutations, gene translocation, gene amplifications and aberrant gene methylation are all reported in papillary thyroid cancers (10). Identification of high-risk PTC patients is beneficial for individualized treatment and surveillance. However, the molecular characteristics of high-risk thyroid cancer remained unclear (11).
Rap1GAP is a GTPase-activating protein that inactivates Rap1-GTP, which is the functional form of Rap1. Rap1GAP has been identified to be suppressed in colon cancer (12,13), pancreatic cancer (14), and thyroid cancer (15). In patients with gastric cancer, lower Rap1GAP expression was associated with larger tumor size, advanced tumor-node-metastasis (TNM) stage and worse prognosis (16). In vitro and in vivo assays revealed that loss of Rap1GAP promoted pancreatic cancer cell growth, survival, and invasion (14). Mitogen-activated protein kinase (MAPK) signaling pathway is reported to be its possible downstream pathway to regulate cell invasion and tumor metastases (17).
The mechanism of Rap1GAP suppression has been investigated in a few previous works. Genetic variation and epigenetic modification both participate to decrease Rap1GAP mRNA and protein level (18). However, there is no research that directly focuses on the relationship between Rap1GAP and miRNAs. Recent literature indicates that miRNAs are essential in thyroid cancer diagnosis, treatment, and prognosis (19,20). These short, non-coding RNAs contribute to regulating the expression of protein-coding genes through binding to 3' untranslated regions (3'UTR) of a gene, resulting in the inhibition of protein translation.
In this article, we defined Rap1GAP protein as a diagnostic marker of PTC and proposed a new mechanism for Rap1GAP protein with miR-3121-3p. MiR-3121-3p showed its potential to suppress Rap1GAP expression and promote PTC cell migration and proliferation via Rap1GAP and MAPK signaling pathway. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4469).
Methods
Database and statistical analysis
The data of 580 papillary thyroid carcinoma samples with mRNA expression matrix and clinical information from The Cancer Genome Atlas (TCGA) were downloaded via cBioportal (https://www.cbioportal.org/) and UCSC (https://xenabrowser.net/). R 3.6.0 was employed for analysis of data from TCGA. The expression of Rap1GAP in tumor and adjacent tissues or paired adjacent tissues were compared using RNA sequencing data. Unpaired or paired t-test was used in comparing Rap1GAP expression in tumors and adjacent tissues or paired adjacent tissues. Receiver operating characteristic (ROC) curve and Wilcoxon signed-rank test were used to evaluate the diagnostic power of Rap1GAP expression in PTC. R package pROC was applied to calculate the best cutoff value for PTC diagnosis and estimate the sensitivity and specificity. A heatmap was drawn to demonstrate the relationship between clinical characteristics and Rap1GAP expression using R package pheatmap. Chi-square was used when analyzing Rap1GAP expressions grouped by clinical features. A P value <0.05 (two tailed) was considered statistically significant.
The experiment data were analyzed via GraphPad Prism 7.0 with unpaired t-tests, and mean value ± standard deviation (SD) were employed to present the results. All in vitro experiments and assays were repeated at least three times. A P value <0.05 (two tailed) was considered statistically significant.
Prediction of Rap1GAP-related miRNAs
TargetScan (21), miRDB (22), mirDIP (23), and microT-CDS in Diana Tools (24) were used to estimate miRNAs paired with 3'UTR of Rap1GAP. These databases created different algorithms to predict latent miRNA-mRNA pairs. The lists of miRNAs were overlapped and those miRNAs predicted in more than three databases with higher match scores were selected as our targets. The binding site between miRNAs and Rap1GAP was predicted in TargetScan, miRDB, and Diana Web Tool.
Cell culture
Human PTC cell lines TPC-1 (RRID: CVCL_6298), B-CPAP (DSMZ Cat# ACC-273, RRID: CVCL_0153) and human embryonic kidney cell line HEK293T (RRID: CVCL_0063) cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences (CAS). Human PTC cell line K1 (RRID: CVCL_2537) cell line was obtained from Guangzhou Cellcook Biotech Co. All cell lines were authenticated by unique short tandem repeats (STRs) reported in the Leibniz Institute DSMZ. TPC-1 and B-CPAP cells were cultured in RPMI-1640 (RPMI 1640 Medium, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco), while K1 cells and 293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (High Glucose, Gibco, USA) supplemented with 10% FBS. All of the cell lines were incubated in humidified 37 °C conditions with 5% CO2.
Cell transfection
Overexpression plasmids of GV-Rap1GAP and negative control vector were provided from GeneChem (Shanghai, China). Small interfering RNAs for human Rap1GAP protein (si-Rap1GAP), mimics and inhibitors of hsa-miR-3121-3p, and corresponding negative controls (si-NC, mimic-NC and inhibitor-NC) were purchased from RiboBio (Guangzhou, China). The dual-luciferase reporter gene vectors were constructed and purchased from RiboBio (Guangzhou, China). Opti-MEM (Gibco, USA) and Lipofectamine 3000 (Invitrogen, USA) were used during transfection according to the manufacturer’s protocol. The medium was changed after 24 hours of transfection.
Dual-luciferase reporter gene assay
Based on bioinformatic prediction, the binding site of miRNAs and 3'UTR of Rap1GAP was selected as a candidate target. The wild-type reporter h-RAP1GAP-WT included the segment of sequence NM_002885.4 from 2245 to 3322 in a total of 1077 bps, and the mutation reporter h-RAP1GAP-MUT was mutated at 3304 to 3310 bp from sequence TCTATTT to AGATAAA. The wild-type reporter h-Rap1GAP-WT or the mutant reporter h-Rap1GAP-MUT was cotransfected with miRNA mimic or miR-control in 293T cells in the following manner. After transfection for 48 hours, Dual-Glo Luciferase Assay System (Promega, WI, USA) was used to assess Firefly luciferase activity normalized to Renilla luciferase activity. Each experiment was repeated three times.
RNA isolation, real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from thyroid cells using RNAiso Plus (TaKaRa, Kyoto, Japan). Reverse transcription of total mRNAs and miR-3121-3p were performed using the PrimeScript™ RT reagent Kit (TaKaRa, Kyoto, Japan). Real time qRT-PCR was performed with the SYBR® Premix Ex Taq™ (Tli RNaseH Plus) (TaKaRa, Kyoto, Japan) and Bio-Rad CFX96 system. MiRNA and mRNA expressions were quantitated and calculated via the method of 2−ΔΔCT, during which U6 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used for normalization, respectively. All reactions were performed in triplicate. Primers and small interfering RNA of Rap1GAP are displayed in Table S1. Reverse transcription and PCR primers of miR-3121-3p and U6 were designed and provided by RiboBio (Guangzhou, China).
Full table
Protein extraction and western blotting
Cells were lysed with RIPA lysis buffer (Servicebio, Wuhan, China) with 1 nM phenylmethylsulfonyl fluoride (PMSF) (Cell Signaling Technology Inc.). Then, the supernatant was boiled with 5*loading buffer (Thermo Fisher, Boston, MA, USA). After electrophoresis and electrotransfer, target proteins were probed to the following primary antibodies: anti-Rap1GAP (1:1,000, Abcam, USA, Cat# ab32373, RRID: AB_777621), anti-Erk1/2 (1:1,000, Cell Signaling Technology, USA, Cat# 9102, RRID: AB_330744), anti-phospho-Erk1/2 (1:1,000, Cell Signaling Technology, USA, Cat# 4695, RRID: AB_390779), anti-Akt (pan) (1:1,000, Cell Signaling Technology, USA, Cat# 4685, RRID: AB_2225340), and anti-phospho-Akt (1:1,000, Cell Signaling Technology, USA, Cat# 4060, RRID: AB_2315049). Anti-Rabbit IgG with horseradish peroxidase (HRP)-linked antibody (1:3,000, Cell Signaling Technology, USA, Cat# 7074, RRID:AB_2099233) was used as the secondary antibody. The visualization steps were described previously (25).
Transwell migration and invasion assay
The migration and invasion abilities of PTC cells were evaluated using Transwell assay (Corning-Costar; pore size 8 µm). Matrigel (Sigma, 5 mg/mL) was pre-coated in chambers after diluted to 1 or 0.6 µg/µL when invasion assay was conducted, and 2×104 cells were planted into one chamber 48 hours after transfection. Cells that moved to the bottom of the membrane were fixed with 4% formaldehyde and stained with 0.5% crystal violet after 24 hours in the migration assay, or after 48 hours in the invasion assay. Each experiment was repeated at least three times. Pictures of cells were taken by inverted microscope (Olympus, Japan) and analyzed using ImageJ v1.52.
Wounding healing assay
For the wound healing assay, 4×105 cells were plated uniformly into 12-well plates 48 hours after the transfection. After the cell attachment, straight lines were drawn through the center of every well. The media with 2% FBS were changed every day. Pictures of the cell migration were taken at 0, 24, and 48 hours after the scratch by inverted microscope (Olympus, Japan) and analyzed using ImageJ v1.52.
Cell Counting-Kit 8 Assay (CCK-8 assay)
A total of 1,000 cells were seeded into 96-well plates with 5 replicates. The appropriate medium with FBS was administered, and the plates were incubated in a 37 °C, 5% CO2 incubator. CCK-8 assay was performed according to the manufacturer’s instruction (Dojindo, Japan) at 24, 48, 72, 96, and 120 hours. The spectrometric absorbance at 450 nm was measured on a microplate reader (Thermo Fisher, USA).
Results
Rap1GAP was a biomarker for PTC diagnosis
Comparing the mRNA expressions between thyroid cancer and unpaired or paired adjacent normal tissues, we found that Rap1GAP was significantly suppressed in tumor tissues (Figure 1A,B, P<0.001). A ROC curve was built, and the area under curve (AUC) was 0.91 (Figure 1C, 95% CI: 0.87–0.94, P<0.001). With a best cutoff value set as the third quartile of Rap1GAP expression, the corresponding sensitivity and specificity were 96.61% (95% CI: 88.29–99.59%) and 83.76% (95% CI: 80.25–86.87%), respectively. BRAFV600E mutation, as a well-established molecular marker for PTC diagnosis, possesses a perfect specificity but limited sensitivity (0–83%) depending on the frequency of gene mutation and detection procedures (26). These results indicated the putative function of Rap1GAP to further improve PTC diagnostic sensitivity.
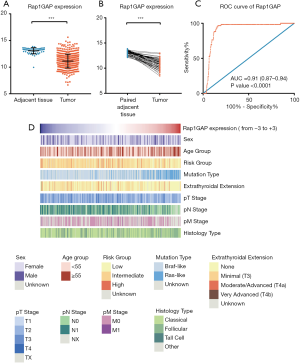
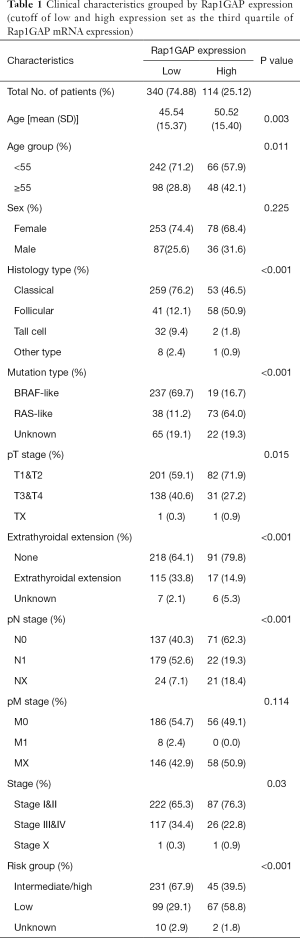
The heatmap illustrated clinical characters including age, sex, histology subtype, mutation group (BRAF mutation vs. RAS mutation), extrathyroidal extension, TNM stage (according to the The American Joint Committee on Cancer (AJCC) 6th or 7th version based on the collection time) and recurrence risk strategy. We divided samples into low expression and high expression groups (340 vs. 114 cases) among 454 cases with relatively complete clinical materials according to Rap1GAP mRNA. Interestingly, we found younger age (P=0.003), BRAF mutation rather than RAS mutation (P<0.001), classical subtype (P<0.001), extrathyroidal extension (P<0.001), advanced T stage (P=0.015), advanced N stage (P<0.001), and high or intermediate risk grouping (P<0.001) were associated with low expression of Rap1GAP mRNA (Figure 1D and Table 1).
Full table
MiR-3121-3p was predicted and confirmed to bind to and suppress Rap1GAP
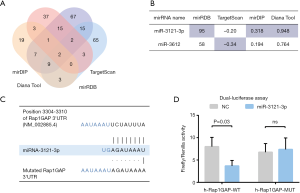
We searched TargetScan, miRDB, mirDIP, and Diana Web Tool for potential miRNAs binding to 3'UTR of Rap1GAP. Subsequently, 64, 44, 118, and 99 miRNAs were found in the website tools above, respectively. There were two miRNAs (miR-3121-3p and miR-3612) that remained after overlapping these results (Figure 2A). We compared their predictive scores in these tools. MiRNA-3121-3p was picked as our further object of research (Figure 2B) since it scored higher in three databases while miR-3612 did so in only one. As displayed in Figure 2C, miRNA-3121-3p has a fragment of 3'-AGAUAAAU-5' which is accurately paired with RAP1GAP 3'UTR 5'-UCUAUUUA-3'. Based on bioinformatic analysis, we conducted dual-luciferase reporter assay to confirm the binding and function of miR-3121-3p. As illustrated in Figure 2D, after transfection of miR-3121-3p mimic, the activation of wild-type Rap1GAP promoter was significantly suppressed while mutated Rap1GAP promoter was not, which indicated that miR-3121-3p could bind to a specific fragment of 3'UTR of Rap1GAP promoter and suppress transcription.
Rap1GAP mRNA expression was significantly suppressed, which coordinated with miR-3121-3p mimic treatment (Figure 3A), and was significantly upregulated after miR-3121-3p inhibition (Figure 3B). The Rap1GAP protein levels showed similar trends as mRNA changes in each group (Figure 3C). Erk kinase was phosphorylated with miR-3121-3p mimic and this effect was reversed by Rap1GAP overexpression. Meanwhile, AKT phosphorylation was similar in these groups, indicating that MAPK/ERK signaling pathway, instead of PI3K/AKT, may be a downstream target of Rap1GAP.
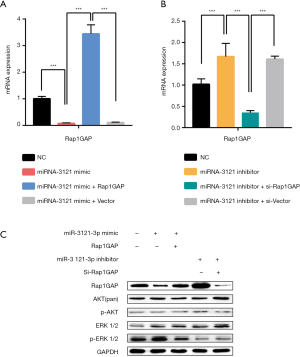
MiR-3121-3p promotes cell migration and invasion via Rap1GAP
Upregulating miRNA-3121-3p in three PTC cells (TPC-1, BCPAP, and K1) enhanced cell migration as demonstrated in Figure 4A,B,C,D. The ability of BCPAP cell invasion was also improved after miRNA-3121-3p mimic treatment (Figure 4A). The ability of upregulated miR-3121-3p cells to migrate and invade was nullified after Rap1GAP overexpression (Figure 4A,B).
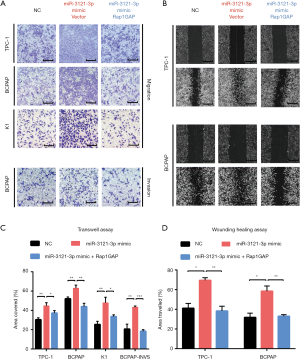
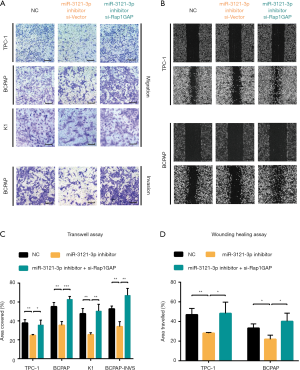
Moreover, we investigated the effect of silencing miR-3121-3p by miRNA inhibitor. MiR-3121-3p inhibitor administration led to less cell migration and invasive properties in Transwell assay (Figure 5A) and wound healing assay (Figure 5B). The statistical analyses showed the significance (Figure 5C,D). Furthermore, silencing Rap1GAP could restore the effects of miR-3121-3p inhibitor in these cells (Figure 5A,B,C,D).
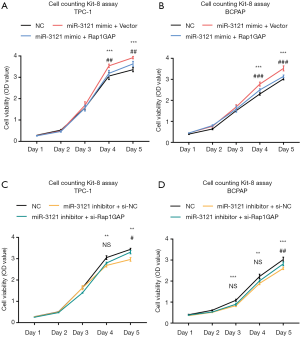
MiR-3121-3p induced cell proliferation through Rap1GAP
MiR-3121-3p mimic treatment enhanced cell proliferation of TPC-1 and BCPAP (Figure 6A,B), while miR-3121-3p inhibitor suppressed proliferation in these cells (Figure 6C,D). The influence of miR-3121-3p mimic and inhibitor on cell growth could partially be reversed by Rap1GAP overexpression or siRNA-mediated knockdown, respectively. We assumed that there was also other unknown mechanisms regulating cell growth induced by miR-3121-3p.
Discussion
In this study, we originally reported the regulation between miR-3121-3p and Rap1GAP and their potential clinical impacts in human cancers. There are very few studies that have focused on the expression and potential functions of miR-3121-3p in human malignancies. MiR-3121-3p was reported to be upregulated in bone marrow in children with acute lymphoblastic leukemia (27), miR-3121-3p overexpression was found in the urine extracellular vesicles of lung cancer patients (28), while miR-3121-3p expression was revealed to be elevated after administration of trastuzumab in breast cancer cells (29). Overall, limited evidence suggests that miR-3121-3p expression has ambiguous promoting effects in human malignancies. However, no studies have examined the role of miRNA in PTC or other thyroid malignancies.
A few studies reported the potential patterns by which miR-3121-3p may influence biological processes. Induced by BCR-ABL tyrosine kinase inhibitors, miR-3121-3p was declined in chronic myeloid leukemia and IL-1β and adhesion molecule expressions were elevated (30). Chen and colleagues detected germline DNA variants and identified novel single nucleotide polymorphisms that could possibly be paired with miR-3121-3p and could lead to tumorigenesis, proliferation, and metastasis (31). Microsatellite instability and CpG island methylator phenotype were also related to miR-3121-3p in colorectal cancer (32). Overall, miR-3121-3p has shown its potential to be a tumor modulator, but there remains questions concerning its clinical significance and mechanisms in malignancies, particularly thyroid cancer. While there is still not a sufficient amount of expression data to confirm the clinical impact miRNA, this study provides in vitro evidence of that miR-3121-3p enhances cell proliferation and spreading in PTC via inhibiting Rap1GAP.
A few previous studies have reported on the mechanisms for Rap1GAP suppression other than those mediated by miRNAs. Genetic variations and epigenetic modifications might be determinants of Rap1GAP expression. In one study comprising 197 cases, Rap1GAP copy number loss was reported in 41% of PTC patients, and the genetic polymorphism of Rap1GAP, Y609C, was seen in 7% of thyroid tumors, but this polymorphism did not lead to Rap1GAP expression change (18). Rap1GAP is known to be a tumor suppressor and its copy number loss was found in pancreatic cancer (14). Moreover, hypermethylation of the promoter and histone deacetylation of Rap1GAP were related to Rap1GAP expression in melanoma (33) and anaplastic thyroid cancer (34). Similarly, Rap1GAP promoter hypomethylation was reported to upregulate Rap1GAP in myelodysplastic syndrome (35). Moreover, Rap1GAP was also shown to be dynamically regulated by thyroid-stimulating hormone (TSH) through phosphorylation followed by proteasome-mediated degradation (36,37). In conclusion, miR-3121-3p, together with hypermethylation, histone deacetylation and protein phosphorylation, have been found to play pivotal roles in modulating Rap1GAP expression.
In our study, we found Rap1GAP expression was correlated to BRAFV600E mutation, extrathyroidal extension, and lymph node metastasis, indicating Rap1GAP’s importance in PTC aggressiveness. Loss of Rap1GAP also contributed to aggressiveness in other cancers. With decreased mRNA and protein levels in gastric cancer, Rap1GAP was found to be an independent prognostic factor and to be significantly associated with tumor size, histological grade, and clinical stage (38). As a GTPase-activating protein of Rap1, Rap1GAP provides a catalytic asparagine to stimulate GTP hydrolysis and subsequently represses the mechanism of Rap1 activity (39). As Rap1 has been well demonstrated to be a BRAF activator in previous studies (40,41), Rap1GAP loss may lead to overactivation of Rap1 and downstream BRAF and MAPK signaling pathway. In previous literature, the MAPK signaling pathway and the PI3K/AKT signaling pathway are activated in papillary thyroid cancer and follicular thyroid cancer, respectively(10). Other researchers also summarized the interaction of MAPK signaling pathway and PI3K/AKT pathways in resistance to radioactive iodine-therapy (42), tumorigenesis (43) and tumor development (44). The MAPK signaling pathway is generally overactivated in PTC with BRAFV600E mutation and plays a detrimental role in tumorigenesis and progression while PI3K/AKT pathway is related to several aberrant receptor tyrosine kinases and results in activation of mammalian target of rapamycin (mTOR) or forkhead box O (FOXO) (45). Our results also confirmed the regulation between miR-3121-3p, Rap1GAP, and ERK phosphorylation in PTC cell lines but not the AKT signaling pathway. There are other mechanisms by which Rap1GAP provokes tumor aggressiveness. In previous studies, Rap1GAP was reported to be correlated to epithelial-mesenchymal transition (EMT) markers including E-cadherin, MMP-2, and MMP-9 (14,16). In colon cancer, Rap1GAP could modulate cell-cell and cell-matrix adhesion, which is an early step of tumor metastasis (12). Rap1GAP also impaired the ability of cells to spread and migrate in collagen, and Focal adhesion kinase (FAK) and steroid receptor coactivator (Src) were identified as downstream signals of Rap1GAP depletion capable of regulating cellular architecture and motility (46).
In conclusion, our results indicate that Rap1GAP is a diagnostic marker in PTC and associated with various clinicopathological features. MiR-3121-3p, as predicted, was proven to bind to Rap1GAP promoter, suppress protein expression, and further promote PTC cell migration, invasion, and proliferation. Elevated Rap1GAP could counteract the aggressiveness induced by miR-3121-3p, with MAPK signaling pathway potentially being involved in these effects. Future works should clarify the clinical significance of miR-3121-3p with sufficient miRNA expression data in PTCs. The potential of therapeutically targeting Rap1GAP and associated miRNAs should also be more comprehensively discussed.
Acknowledgments
We would like to thank all the colleagues who helped us during all the subject design and experimental study.
Funding: This work was supported by Hubei Provincial Natural Science Foundation of China grant 2015CKB733 to Pengfei Yi.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4469
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4469
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4469). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Xing M, Alzahrani AS, Carson KA, et al. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J Clin Oncol 2015;33:42-50. [Crossref] [PubMed]
- Tuttle RM, Tala H, Shah J, et al. Estimating risk of recurrence in differentiated thyroid cancer after total thyroidectomy and radioactive iodine remnant ablation: using response to therapy variables to modify the initial risk estimates predicted by the new American Thyroid Association staging system. Thyroid 2010;20:1341-9. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014;159:676-90. [PubMed]
- Xing M, Westra WH, Tufano RP, et al. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. J Clin Endocrinol Metab 2005;90:6373-9. [Crossref] [PubMed]
- Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocr Rev 2007;28:742-62. [Crossref] [PubMed]
- Kim TH, Park YJ, Lim JA, et al. The association of the BRAF(V600E) mutation with prognostic factors and poor clinical outcome in papillary thyroid cancer: a meta-analysis. Cancer 2012;118:1764-73. [Crossref] [PubMed]
- Liu R, Xing M. TERT promoter mutations in thyroid cancer. Endocr Relat Cancer 2016;23:R143-55. [Crossref] [PubMed]
- Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer 2013;13:184-99. [Crossref] [PubMed]
- Yip L, Sosa JA. Molecular-Directed Treatment of Differentiated Thyroid Cancer: Advances in Diagnosis and Treatment. JAMA Surg 2016;151:663-70. [Crossref] [PubMed]
- Tsygankova OM, Ma C, Tang W, et al. Downregulation of Rap1GAP in human tumor cells alters cell/matrix and cell/cell adhesion. Mol Cell Biol 2010;30:3262-74. [Crossref] [PubMed]
- Gao WL, Ye GC, Liu LW, et al. The downregulation of Rap1 GTPase-activating protein is associated with a poor prognosis in colorectal cancer and may impact on tumor progression. Oncol Lett 2018;15:7661-8. [Crossref] [PubMed]
- Zhang L, Chenwei L, Mahmood R, et al. Identification of a putative tumor suppressor gene Rap1GAP in pancreatic cancer. Cancer Res 2006;66:898-906. [Crossref] [PubMed]
- Nellore A, Paziana K, Ma C, et al. Loss of Rap1GAP in papillary thyroid cancer. J Clin Endocrinol Metab 2009;94:1026-32. [Crossref] [PubMed]
- Yang Y, Zhang J, Yan Y, et al. Low expression of Rap1GAP is associated with epithelial-mesenchymal transition (EMT) and poor prognosis in gastric cancer. Oncotarget 2017;8:8057-68. [Crossref] [PubMed]
- Mochizuki N, Ohba Y, Kiyokawa E, et al. Activation of the ERK/MAPK pathway by an isoform of rap1GAP associated with G alpha(i). Nature 1999;400:891-4. [PubMed]
- Zuo H, Gandhi M, Edreira MM, et al. Downregulation of Rap1GAP through epigenetic silencing and loss of heterozygosity promotes invasion and progression of thyroid tumors. Cancer Res 2010;70:1389-97. [Crossref] [PubMed]
- Wojcicka A, Kolanowska M, Jazdzewski K. MicroRNA in diagnostics and therapy of thyroid cancer. Eur J Endocrinol 2016;174:R89-98. [Crossref] [PubMed]
- Lee JC, Gundara JS, Glover A, et al. MicroRNA expression profiles in the management of papillary thyroid cancer. Oncologist 2014;19:1141-7. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [Crossref] [PubMed]
- Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol 2019;20:18. [Crossref] [PubMed]
- Tokar T, Pastrello C, Rossos AEM, et al. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res 2018;46:D360-D70. [Crossref] [PubMed]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 2013;41:W169-73. [Crossref] [PubMed]
- Maimaiti Y, Jie T, Jing Z, et al. Aurora kinase A induces papillary thyroid cancer lymph node metastasis by promoting cofilin-1 activity. Biochem Biophys Res Commun 2016;473:212-8. [Crossref] [PubMed]
- de Koster EJ, de Geus-Oei LF, Dekkers OM, et al. Diagnostic Utility of Molecular and Imaging Biomarkers in Cytological Indeterminate Thyroid Nodules. Endocr Rev 2018;39:154-91. [Crossref] [PubMed]
- Duyu M, Durmaz B, Gunduz C, et al. Prospective evaluation of whole genome microRNA expression profiling in childhood acute lymphoblastic leukemia. Biomed Res Int 2014;2014:967585. [Crossref] [PubMed]
- Yasui T, Yanagida T, Ito S, et al. Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci Adv 2017;3:e1701133. [Crossref] [PubMed]
- Cilek EE, Ozturk H, Gur Dedeoglu B. Construction of miRNA-miRNA networks revealing the complexity of miRNA-mediated mechanisms in trastuzumab treated breast cancer cell lines. PLoS One 2017;12:e0185558. [Crossref] [PubMed]
- Sukegawa M, Wang X, Nishioka C, et al. The BCR/ABL tyrosine kinase inhibitor, nilotinib, stimulates expression of IL-1beta in vascular endothelium in association with downregulation of miR-3p. Leuk Res 2017;58:83-90. [Crossref] [PubMed]
- Chen X, Paranjape T, Stahlhut C, et al. Targeted resequencing of the microRNAome and 3'UTRome reveals functional germline DNA variants with altered prevalence in epithelial ovarian cancer. Oncogene 2015;34:2125-37. [Crossref] [PubMed]
- Mullany LE, Herrick JS, Wolff RK, et al. Association of cigarette smoking and microRNA expression in rectal cancer: Insight into tumor phenotype. Cancer Epidemiol 2016;45:98-107. [Crossref] [PubMed]
- Zheng H, Gao L, Feng Y, et al. Down-regulation of Rap1GAP via promoter hypermethylation promotes melanoma cell proliferation, survival, and migration. Cancer Res 2009;69:449-57. [Crossref] [PubMed]
- Dong X, Korch C, Meinkoth JL. Histone deacetylase inhibitors upregulate Rap1GAP and inhibit Rap activity in thyroid tumor cells. Endocr Relat Cancer 2011;18:301-10. [Crossref] [PubMed]
- Ding WJ, Yang Y, Chen ZX, et al. Methylation level of Rap1GAP and the clinical significance in MDS. Oncol Lett 2018;16:7287-94. [Crossref] [PubMed]
- Tsygankova OM, Feshchenko E, Klein PS, et al. Thyroid-stimulating hormone/cAMP and glycogen synthase kinase 3beta elicit opposing effects on Rap1GAP stability. J Biol Chem 2004;279:5501-7. [Crossref] [PubMed]
- Rubinfeld B, Crosier WJ, Albert I, et al. Localization of the rap1GAP catalytic domain and sites of phosphorylation by mutational analysis. Mol Cell Biol 1992;12:4634-42. [Crossref] [PubMed]
- Zhao J, Mai C, Weng D, et al. Reduced expression of Rap1GAP as a prognostic biomarker for primary gastric cancer patients. Cancer Biomark 2018;22:375-84. [Crossref] [PubMed]
- Daumke O, Weyand M, Chakrabarti PP, et al. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature 2004;429:197-201. [Crossref] [PubMed]
- Dorard C, Cseh B, Ehrenreiter K, et al. RAF dimers control vascular permeability and cytoskeletal rearrangements at endothelial cell-cell junctions. FEBS J 2019;286:2277-94. [Crossref] [PubMed]
- Garcia J, de Gunzburg J, Eychene A, et al. Thrombopoietin-mediated sustained activation of extracellular signal-regulated kinase in UT7-Mpl cells requires both Ras-Raf-1- and Rap1-B-Raf-dependent pathways. Mol Cell Biol 2001;21:2659-70. [Crossref] [PubMed]
- Liu J, Liu Y, Lin Y, et al. Radioactive Iodine-Refractory Differentiated Thyroid Cancer and Redifferentiation Therapy. Endocrinol Metab (Seoul) 2019;34:215-25. [Crossref] [PubMed]
- Zaballos MA, Santisteban P. Key signaling pathways in thyroid cancer. J Endocrinol 2017;235:R43-R61. [Crossref] [PubMed]
- Jin S, Borkhuu O, Bao W, et al. Signaling Pathways in Thyroid Cancer and Their Therapeutic Implications. J Clin Med Res 2016;8:284-96. [Crossref] [PubMed]
- Nozhat Z, Hedayati M. PI3K/AKT Pathway and Its Mediators in Thyroid Carcinomas. Mol Diagn Ther 2016;20:13-26. [Crossref] [PubMed]
- Dong X, Tang W, Stopenski S, et al. RAP1GAP inhibits cytoskeletal remodeling and motility in thyroid cancer cells. Endocr Relat Cancer 2012;19:575-88. [Crossref] [PubMed]


