
Gene set enrichment analysis and protein-protein interaction network analysis after sciatic nerve injury
Introduction
Peripheral nerve injuries (PNIs) present a substantial clinical problem worldwide and are associated with a considerable financial burden (1). Direct suturing repair, commonly used for short nerve deficits (<0.5 cm), is a gold-standard treatment for axonotmesis and neurotmesis injuries (2). A repair of larger nerve deficits requires nerve autografting, which means implanting a patient’s healthy nerves (3). With the limitation of nerve autografting, peripheral nerve regeneration aided by tissue engineering technology emerged as the alternatives (4). Owing to the different cell types in the peripheral nervous system (PNS) and the central nervous system (CNS), and their specific functions post-injury, peripheral nerves possess greater potential for regeneration than those in the CNS (5). Due to the traumatic injury and postoperative complications, PNIs can trigger a series of biological events within the proximal and distal nerve stumps (6). Substantial progress has been made with understanding how PNIs elicit these transcriptional and epigenetic changes (7). Regulators of axonal regeneration have also been identified, which has provided valuable insight into understanding the transcriptional changes that promote regeneration in peripheral neuron response to injury (8); however, there are still many questions left to answer.
Beyond the classical injury signal pathways, transcriptional and epigenetic factors have been shown to be involved in axonal regeneration, which can be facilitated by pharmacological or genetic means to conquer the non-permissive microenvironment in vitro or in vivo (9-14). Therefore, a clear understanding of peripheral nerve regeneration mechanisms would allow us to offer feasible treatment for patients with PNIs. Furthermore, it may provide potential clues for research into CNS regeneration.
The dynamic changes of biological processes and related core genes that occurred in both sciatic nerve stumps during neuroregeneration in rats were reported in our previous research (15-18). In particular, the molecular pathways and interactive networks underlying the molecular interaction between immune response and axonal regeneration following sciatic nerve transection were discussed (15). The immune response followed by a persistent hyperinflammatory state accompanied by increased infiltration of macrophages and inflammatory signals has been reported after sciatic nerve injury (19). Macrophages enhance tissue clearance, which is further promoted by cytokine release by CD-4-positive T cells and B lymphocyte-produced antibodies (20). In addition to immune response, chemotaxis (21), blood vessel regeneration (16), and remyelination (22) are crucial events after PNI. To obtain a more comprehensive understanding of the biological processes, cellular components, molecular functions, signaling pathways, and protein-protein interaction (PPI) networks involved in the intrinsic regenerative programs after PNI, more wide-ranging investigations are needed.
To investigate the molecules and pathways involved after PNI systematically, we performed gene set enrichment analysis (GSEA), constructed PPI networks, and verified gene expression ex vitro at 1, 4, 7, and 14 days after sciatic nerve transection in rats. Neural precursor cell expressed developmentally downregulated 4-like (NEDD4L), neuregulin 1 (NRG1), nuclear factor of activated T cells 2 (NFATC2), midline 1 (MID1), GLI family zinc finger 2 (GLI2), and ventral anterior homeobox 1 (VAX1), which were involved in both immune response and axonal regeneration following sciatic nerve transection, were screened. NEDD4L, NRG1, NFATC2, MID1, GLI2, and VAX1 are all related to neural development or other functions in the nervous system (23-37). Of these genes, VAX1, the known functions of which include guidance, binding, and penetration of axons, was considered to be a potential therapeutic target, because its expression continuously increased after nerve injury. Our work showed that the injury-induced molecular change in proximal nerve stump includes enriched molecular pathways and PPI networks. Furthermore, mechanistic-based treatments may potentially be developed based on our work, which can be used as a tool for exploring the potential therapeutic target in neuroregeneration after PNI. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4958).
Methods
Animal model
Adult male Sprague-Dawley (SD) rats (weight: 200–220 g, supplied by the Experimental Animal Center of Nantong University) were randomly divided into five groups (15 rats in each group). The rats were anesthetized by intraperitoneal injection of composite narcotics, consisting of trichloroac etaldehyde monohydrate (85 mg/kg), magnesium sulfate (42 mg/kg), and sodium pentobarbital (17 mg/kg). Then, an incision was made on the lateral aspect of the left mid-thigh of the rats, and the sciatic nerve was identified. The sciatic nerve transection was carried out near the center of the femur, and the incision was closed. All animal experiments in this study were performed in accordance with the guidelines for animal care and were approved by the Administration Committee of Experimental Animals, Jiangsu Province, China [SYXK (Su) 2012-0031].
Sample and raw data collection
All samples were collected according to the previous protocol (38). Briefly, 0.5 cm of non-injured nerves and the proximal sciatic nerve stumps were collected at 1, 4, 7, 14 days post injury, respectively. Five groups were divided by the time points preoperatively and postoperatively (normal, 1, 4, 7, and 14 days). Total RNA was extracted from the nerve samples using Trizol (Life technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The RNA quality of each sample was qualified using Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, USA) and Nanodrop ND1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Microarray analysis was performed with an Agilent Microarray Scanner (Agilent Technologies), and the subsequent data were compiled with Agilent feature extraction software. All steps from RNA amplification to the final scanner output were conducted by the National Engineering Center for Biochip at Shanghai (China), and three biological replicates were performed for each group. The raw data can be accessed from the Gene Expression Omnibus (GEO) database (GSE30165).
Bioinformatics analysis
Log2-transformed mean-centered datasets filtered for expression values greater than 128 in any subsets and including only 10% of probes were used for further analysis (15). GSEA (Broad Institute; software.broadinstitute.org/gsea/) was performed to show variation among groups, with probes ranked by signal-to-noise ratio and statistical significance determined by 1,000 gene set permutations (39). For GSEA analysis, a GO gene set enrichment map generated by a false discovery rate (FDR). Q-value cutoff of 0.01 was used to obtain credible results. The results of gene set enrichment were graphically mapped to the Enrichment Map in Cytoscape (US National Institute of General Medical Sciences, Bethesda, MD, USA). Node size represents the number of genes in the gene set, and edge thickness is proportional to the overlap between gene sets, calculated using overlap coefficients. The enrichment score was mapped to the node color as a color gradient (40). The Venn diagrams were created using the Venny 2.1.0 online tool (41). For genes with fold change (FC) ±2.0, pathway analysis and the construction of PPI networks of the expression data were performed with Ingenuity Pathway Analysis (IPA, QIAGEN, Redwood City, CA, USA). Pathways with Z-score ≥2 or ≤−2 (Benjamini-Hochberg method) were considered to be significant. PPI networks were depicted on the IPA database.
Quantitative real-time polymerase chain reaction (qPCR)
Reverse-transcribed complementary DNA synthesis and qPCR were sequentially performed with the Prime-Script RT reagent Kit (TaKaRa, Dalian, China) and SYBR Premix Ex Taq (TaKaRa, Dalian, China), respectively. The relative expression levels of genes were calculated by comparative 2−ΔΔCt method. The sequences of primer pairs used are provided in Table S1.
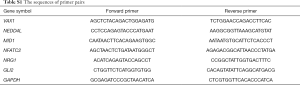
Full table
Histological immunofluorescent staining
At 1, 4, 7 and 14 days after surgery, nerve samples from the proximal nerve stumps and normal nerve sections (0 d) of the rats were harvested and cut into longitudinal sections. The samples were then subjected to immunofluorescent triple-staining with rabbit anti-VAX1 (1:400 dilution, Sigma), mouse anti-NF200 (1:400 dilution, Sigma), and Hoechst 33342 (1:5,000 dilution, Life Technologies) respectively. The nerve sections were incubated with primary antibody at 4 °C overnight, followed by further incubation with the secondary antibody (Goat anti-Mouse IgG-Alex-488, 1:1,000 and Donkey anti-Rabbit IgG-Cy3, 1:1,000) at 4 °C overnight. Finally, the nerve sections were observed under a confocal laser scanning microscope (TCS SP2, Leica).
Statistical analysis
For statistical analysis, the data were replicated in at least three independent experiments. Data are showed as mean ± standard error of the mean (SEM). Multiple comparisons were performed with one-way analysis of variance (ANOVA) as well as Bonferroni post-hoc t-test. The statistical analyses were carried out using IBM SPSS Statistics 26.0 (IBM Corp., Armonk, NY, USA). Differences were considered significant at *P value <0.05, and **P value <0.01.
Results
Overview of functional GSEA in rat proximal nerve stumps following sciatic nerve transection
To elucidate the mechanism of neuroregeneration after sciatic nerve transection, the microarray data from proximal nerve stumps and normal nerve sections of rats were analyzed by GSEA and IPA at 1, 4, 7, 14 days after surgery. qPCR and immunofluorescent staining were also carried out to confirm the expression and histological localization of the potential genes for immune response and axonal regeneration (Figure 1A).
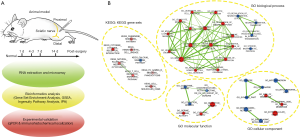
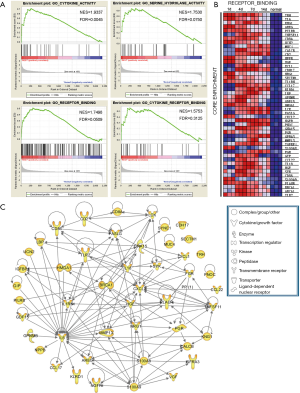
GSEA with enrichment map analysis was performed to visualize the enriched biological processes, cellular components, molecular functions, and KEGG pathways following sciatic nerve transection. The only gene sets passing significance thresholds (FDR Q-value cutoff: 0.01) were selected for inclusion in the Enrichment Map (Figure 1B). The enrichment data showed that the gene sets of GO biological processes related to immune response, chemotaxis, cell activation, and cell adhesion were upregulated in the nerve samples after injury. In contrast, in the normal nerve samples (control group), the neurotransmitter-related biological processes and steroid synthesis were upregulated. For GO cellular components, gene sets enriched in extracellular space and cell membrane were upregulated after injury, while the synapse, mitochondria, and endoplasmic reticulum were upregulated in the control group. For GO molecular functions, gene sets enriched in receptor binding, cytokine activity, and serine hydrolase activity were upregulated after injury, while co-enzyme and con-factor binding were upregulated in the control group. In KEGG analysis, the most upregulated gene sets after injury were involved in immune response, cytokine and cell adhesion-related signaling pathways, while only the calcium signaling pathway was upregulated in the control group. All of the above GSEA data indicated that when injured, the sciatic nerve exhibited an intensive immune response, which may have activated the regeneration process; this is consistent with the findings of previous reports (15). Due to the direct impact of mechanical injury, the neurofunctional gene sets and signaling pathways were inhibited, but this was not our main focus.
GSEA of critical GO biological processes after sciatic nerve transection
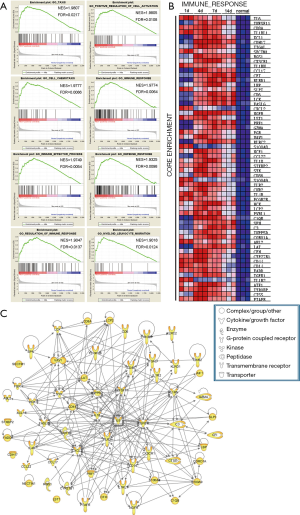
GSEA was performed to determine the enriched gene sets of critical GO biological processes after sciatic nerve transfection. After sciatic nerve transection, the top 8 upregulated gene sets according to normalized enrichment score (NES) had the highest overlapping genes involved in immune response, defense response, leukocyte migration, cell chemotaxis, and cell activation (Figure 2A). The dynamic expression of the core enrichment genes involved in immune response are shown in a heat map in Figure 2B. The proteins encoded by these genes including enzymes, cytokines, peptidases, G-protein coupled receptors, kinases, transmembrane receptors, and other molecules, were integrated into the PPI network based on the IPA database (Figure 2C). Of these proteins, interleukin 6 (IL-6) and transforming growth factor beta 1 (TGFB1) exhibited higher levels of activity than the others, which suggested that they play crucial roles during neuroregeneration after injury.
GSEA of critical GO cellular components after sciatic nerve transection
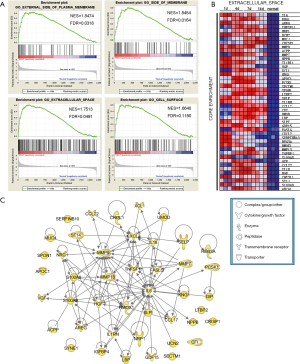
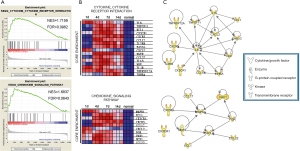
GSEA of GO cellular components showed that the top 4 upregulated gene sets after sciatic nerve transection had the highest overlapping genes localized in the membrane, extracellular space, and cell surface (Figure 3A). The dynamic expression of the core enrichment genes localized in the extracellular space is shown in a heat map in Figure 3B. The proteins encoded by these genes including enzymes, cytokines, peptidases, transporters, and transmembrane receptors, and their interactions are shown in the PPI network displayed in Figure 3C. Besides IL-6, CXCL2 and MMP9 also have been demonstrated to involve in neuroregeneration (42,43).
GSEA of critical GO molecular functions after sciatic nerve transection
GSEA of GO molecular functions showed that the top 4 upregulated gene sets after sciatic nerve transection had the highest overlapping genes involved in cytokine activity, serine hydrolase activity, and receptor binding (Figure 4A). The heat map in Figure 4B shows the dynamic expression of the core enrichment genes involved in receptor binding. A PPI network was constructed integrating the enzymes, cytokines, transcription regulators, peptidases, kinases, transporters, transmembrane receptors, and ligand-depend nuclear receptors encoded by these genes (Figure 4C). Similar to the previous report’s findings, we observed that BRCA1 and TLR2 potentially play essential roles in neuroregeneration (44,45).
GSEA of critical KEGG pathways after sciatic nerve transection
The top 2 enriched KEGG pathways after sciatic nerve transection were the cytokine-cytokine receptor interaction and chemokine signaling pathways (Figure 5A). Figure 5B shows a heat map of the dynamic expression of the core enrichment genes in these pathways. A PPI network was constructed integrating the cytokines, enzymes, kinases, and transmembrane receptors encoded by the genes in these pathways (Figure 5C). Among these proteins, hepatocyte growth factor (HGF), interleukin-18 (IL-18), inhibin subunit beta A (INHBA), and mitogen-activated protein kinase 1 (MAPK1) were highly correlated with peripheral nerve regeneration, which supports the efficacy of our analytical methods (46-49).
Dynamic canonical pathway enrichment analysis after PNI
Using the IPA comparison module, we identified 35 canonical pathways as being significantly enriched (both activation and inhibition, Z-core ≥2 or Z-core ≤−2) at 1, 4, 7, 14 days after sciatic nerve transection (Figure 6). Most of them were associated with immune response. Meanwhile, “PPAR Signaling”, “LXR/RXR Activation”, “Cell Cycle: G2/M DNA Damage Checkpoint Regulation”, “CDK5 Signaling” were inhibited canonical pathways, while others pathways were activated after PNI. Among them, “TREM1 Signaling”, “Role of NFAT in Regulation of the Immune Response”, “PPAR Signaling”, “LXR/RXR Activation”, “IL-6 Signaling”, “IL-17A Signaling in Airway Cells”, “Ephrin B Signaling”, “TNFR1 Signaling”, and “Signaling by Rho Family GTPases” were revealed to be involved in neuroregeneration (50-58).

qPCR and histological validation of potential genes regulating neuroregeneration
The Venn diagram showed that 39 significantly expressed genes were involved in neuroregeneration at 1, 4, 7, 14 days after sciatic nerve transection (Figure 7A). The proteins encoded by these genes play important roles in axonogenesis, guidance, penetration, binding, regeneration, axon growth, outgrowth, branching, and myelination of axons. Interestingly, most of these genes were involved in immune response (Figure 7B). To further confirm the microarray data, six differentially expressed genes including NEDD4L, NRG1, NFATC2, MID1, GLI2, and VAX1, which are associated with immune response and axonal regeneration, were validated by qPCR (Figure 7C). NEDD4L, NFATC2, MID1, and GLI2 shared a similar expression trend, suggesting that they play similar roles following sciatic nerve transection. NEDD4L mediates the ubiquitination of multiple target substrates and plays a critical role in neural development (23), axonal degeneration (24), and neuropathic pain (25). As a member of the nuclear factor of activated T-cell (NFAT) family, NFATC2 is involved in neural development (26), axon growth, synaptic plasticity, and neuronal survival (27). Mid1 is important for normal axonal development through the promotion of axon growth and branch formation (28). Gli2 is required for the initial extension of axons in the mouse spinal cord (29). NRG1 and its neuronal tyrosine kinase receptor ErbB4 are well-known regulators of myelination in the PNS (30-32). They also regulate synaptic transmission in the CNS (33,34), and influence several processes of neurodevelopment (35).
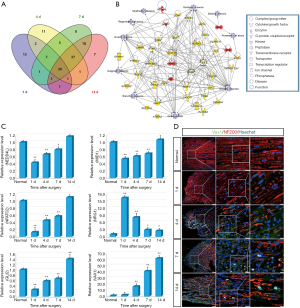
To address the molecular effects of regulating the behavior of neural cells during sciatic nerve regeneration, triple immunostaining of sciatic nerve longitudinal transection was carried out, which validated the dynamic expression level of VAX1 after sciatic nerve transection (Figure 7D).
Discussion
The regeneration of peripheral nerves is often incorrectly understood to occur spontaneously and robustly without improvement or further support. However, injured peripheral nerves rarely recover completely, especially after complicated PNIs (59). Activating neurotrophin pathways, protecting the myelin sheath, and reducing the local response of inflammatory and antioxidative stress, as well as minimizing the scar formation at the lesion site, are the most well-known ways for supporting peripheral nerve regeneration (60). Although the inhibition or knockdown of some tumor suppressors, such as phosphatase and tensin homolog (PTEN) or retinoblastoma 1 (Rb1), may improve axonal regeneration (59), new neuroregenerative targets are needed.
Informed by our previous mRNA profiling data in proximal nerve segments (15,16,38), we jointly applied GSEA, IPA, qPCR, and histological localization to validate the differentially expressed genes at 1, 4, 7, and 14 days after sciatic nerve transection in rats, with the aim of identifying potential therapeutic targets for neuroregeneration. The results of GSEA revealed that molecules located in the extracellular space or cell membrane, as well as signal pathways involved in immune response, chemotaxis, cell activation, and cell adhesion, play crucial roles in neuroregeneration after PNI.
To further investigate the transcriptional dynamics after PNI, canonical pathway enrichment analysis was carried out using IPA, which allowed us to analyze the coordinate expression changes at a pathway level rather than focusing on a single gene. GSEA and IPA obtained similar results. The IL-6, TGFβ, MAPK, and TNF signaling pathways were significantly enriched. The molecules and signal pathways involved in immune response showed unprecedented importance in neuroregeneration.
Furthermore, NEDD4L, NRG1, NFATC2, MID1, GLI2, and VAX1, which potentially play crucial roles after PNI, were selected for follow-up validation. Among them, VAX1 was determined to be a potential therapeutic target for neuroregeneration, because its expression continuously increased after nerve injury. It has also been shown in previous reports to perform the functions of guidance, binding, and penetration of axons (36).
To date, no drug treatment which can improve the speed and quality of peripheral nerve regeneration after PNI has been widely accepted in clinical practice (60). Our data could be used as a tool for screening drugs or small molecular compounds that may enhance axonal regeneration. Technical challenges to navigate the injured axons across gaps between the proximal and distal stump still exist. Unanticipated axon hesitation results in delayed, slow, and staggered neuroregeneration (59). However, our work may lead to the expectation that coordinate overexpression or knockdown of the core set of genes identified here will enhance peripheral nerve regeneration. We predict that changing the transcriptional state of the regenerative microenvironment by coordinate expression of genes involved in immune response may activate the relevant functional molecular pathways. This represents a worthwhile strategy, which may enhance the possibility of creating axonal regenerative capacity after the injury that has been supported by other studies on PNS and CNS (61,62). Because there are no drug sensitivity or targeted therapeutic data currently available, further basic and preclinical work using genetic tools are needed to confirm that Vax1 can serve as a therapeutic target in the treatment of PNIs. Compared to our previous work (15,16,38), this study systematically examined the molecular changes by using both GSEA and PPI network analysis. Immune response and the related signaling pathways were addressed. As a transcriptional regulator, VAX1 was found to be partially localized in the regenerative axons and other cells in the local microenvironment; this needs to be investigated further in the future studies. Axonal proteins possibly transfer from the neuron bodies to the injured axon terminals in response to PNI. VAX1 was previously found to play an essential role in axon guidance, penetration of axons, major tract formation in the developing forebrain (37), and retinal ganglion cell axonal growth (36), which strongly indicates its crucial role in neuroregeneration. VAX1 was screened in our present work by the comprehensive approaches as gene expression profiling followed by multi-level bioinformatics analysis and experimental validation, which were highly correlated with axonal regeneration (Figure 7B). Based on the continually increased expression of VAX1 post-PNI, we hypothesis VAX1 is highly related to the peripheral nerve regeneration. Further experiment as knocking-down/out or over-expressing the expression of these genes in vitro and in vivo should be carried out in order to explore the roles and regular mechanisms of these investigated genes in future.
Conclusions
The combination of GSEA and PPI network analysis served as a valuable tool for identifying the molecular processes and potential therapeutic targets in neuroregeneration.
Acknowledgments
Funding: This work was graciously supported by the National Natural Science Foundation of China (Grant No.81873767), Jiangsu Provincial Key Medical Center and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Technology Project of Nantong (MS12017015-1, JC2018060, JC2019028).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4958
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4958
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4958). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments in this study were performed in accordance with the guidelines for animal care and were approved by the Administration Committee of Experimental Animals [SYXK (Su) 2012-0031] of Jiangsu.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Qiu J, Yang X, Wang L, et al. Isoquercitrin promotes peripheral nerve regeneration through inhibiting oxidative stress following sciatic crush injury in mice. Ann Transl Med 2019;7:680. [Crossref] [PubMed]
- Li R, Liu Z, Pan Y, et al. Peripheral nerve injuries treatment: a systematic review. Cell Biochem Biophys 2014;68:449-54. [Crossref] [PubMed]
- Carvalho CR, Costa JB, Costa L, et al. Enhanced performance of chitosan/keratin membranes with potential application in peripheral nerve repair. Biomater Sci 2019;7:5451-66. [Crossref] [PubMed]
- Zhang PX, Han N, Kou YH, et al. Tissue engineering for the repair of peripheral nerve injury. Neural Regen Res 2019;14:51-8. [Crossref] [PubMed]
- Curcio M, Bradke F. Axon Regeneration in the Central Nervous System: Facing the Challenges from the Inside. Annu Rev Cell Dev Biol 2018;34:495-521. [Crossref] [PubMed]
- Scheib J, Hoke A. Advances in peripheral nerve regeneration. Nat Rev Neurol 2013;9:668-76. [Crossref] [PubMed]
- Mahar M, Cavalli V. Intrinsic mechanisms of neuronal axon regeneration. Nat Rev Neurosci 2018;19:323-37. [Crossref] [PubMed]
- Blackmore MG. Molecular control of axon growth: insights from comparative gene profiling and high-throughput screening. Int Rev Neurobiol 2012;105:39-70. [Crossref] [PubMed]
- Brochier C, Jones JI, Willis DE, et al. Poly(ADP-ribose) polymerase 1 is a novel target to promote axonal regeneration. Proc Natl Acad Sci U S A 2015;112:15220-5. [Crossref] [PubMed]
- Webber C, Zochodne D. The nerve regenerative microenvironment: early behavior and partnership of axons and Schwann cells. Exp Neurol 2010;223:51-9. [Crossref] [PubMed]
- Norsworthy MW, Bei F, Kawaguchi R, et al. Sox11 Expression Promotes Regeneration of Some Retinal Ganglion Cell Types but Kills Others. Neuron 2017;94:1112-20.e4. [Crossref] [PubMed]
- Cho Y, Shin JE, Ewan EE, et al. Activating Injury-Responsive Genes with Hypoxia Enhances Axon Regeneration through Neuronal HIF-1alpha. Neuron 2015;88:720-34. [Crossref] [PubMed]
- Chandran V, Coppola G, Nawabi H, et al. A Systems-Level Analysis of the Peripheral Nerve Intrinsic Axonal Growth Program. Neuron 2016;89:956-70. [Crossref] [PubMed]
- Weng YL, Wang X, An R, et al. Epitranscriptomic m(6)A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 2018;97:313-25.e6. [Crossref] [PubMed]
- Guo Q, Zhu H, Wang H, et al. Transcriptomic Landscapes of Immune Response and Axonal Regeneration by Integrative Analysis of Molecular Pathways and Interactive Networks Post-sciatic Nerve Transection. Front Neurosci 2018;12:457. [Crossref] [PubMed]
- Wang H, Zhu H, Guo Q, et al. Overlapping Mechanisms of Peripheral Nerve Regeneration and Angiogenesis Following Sciatic Nerve Transection. Front Cell Neurosci 2017;11:323. [Crossref] [PubMed]
- Yu J, Gu X, Yi S. Ingenuity Pathway Analysis of Gene Expression Profiles in Distal Nerve Stump following Nerve Injury: Insights into Wallerian Degeneration. Front Cell Neurosci 2016;10:274. [Crossref] [PubMed]
- Yi S, Tang X, Yu J, et al. Microarray and qPCR Analyses of Wallerian Degeneration in Rat Sciatic Nerves. Front Cell Neurosci 2017;11:22. [Crossref] [PubMed]
- Buttner R, Schulz A, Reuter M, et al. Inflammaging impairs peripheral nerve maintenance and regeneration. Aging Cell 2018;17:e12833. [Crossref] [PubMed]
- Bombeiro AL, Santini JC, Thome R, et al. Enhanced Immune Response in Immunodeficient Mice Improves Peripheral Nerve Regeneration Following Axotomy. Front Cell Neurosci 2016;10:151. [Crossref] [PubMed]
- Allard DE, Wang Y, Li JJ, et al. Schwann cell-derived periostin promotes autoimmune peripheral polyneuropathy via macrophage recruitment. J Clin Invest 2018;128:4727-41. [Crossref] [PubMed]
- He X, Zhang L, Queme LF, et al. A histone deacetylase 3-dependent pathway delimits peripheral myelin growth and functional regeneration. Nat Med 2018;24:338-51. [Crossref] [PubMed]
- Rodrigues DC, Mufteev M, Weatheritt RJ, et al. Shifts in Ribosome Engagement Impact Key Gene Sets in Neurodevelopment and Ubiquitination in Rett Syndrome. Cell Rep 2020;30:4179-96.e11. [Crossref] [PubMed]
- Peeters K, Palaima P, Pelayo-Negro AL, et al. Charcot-Marie-Tooth disease type 2G redefined by a novel mutation in LRSAM1. Ann Neurol 2016;80:823-33. [Crossref] [PubMed]
- Laedermann CJ, Cachemaille M, Kirschmann G, et al. Dysregulation of voltage-gated sodium channels by ubiquitin ligase NEDD4-2 in neuropathic pain. J Clin Invest 2013;123:3002-13. [Crossref] [PubMed]
- Graef IA, Wang F, Charron F, et al. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell 2003;113:657-70. [Crossref] [PubMed]
- Vihma H, Luhakooder M, Pruunsild P, et al. Regulation of different human NFAT isoforms by neuronal activity. J Neurochem 2016;137:394-408. [Crossref] [PubMed]
- Lu T, Chen R, Cox TC, et al. X-linked microtubule-associated protein, Mid1, regulates axon development. Proc Natl Acad Sci U S A 2013;110:19131-6. [Crossref] [PubMed]
- Dillon AK, Fujita SC, Matise MP, et al. Molecular control of spinal accessory motor neuron/axon development in the mouse spinal cord. J Neurosci 2005;25:10119-30. [Crossref] [PubMed]
- Ohno M, Hiraoka Y, Matsuoka T, et al. Nardilysin regulates axonal maturation and myelination in the central and peripheral nervous system. Nat Neurosci 2009;12:1506-13. [Crossref] [PubMed]
- Clark AJ, Kaller MS, Galino J, et al. Co-cultures with stem cell-derived human sensory neurons reveal regulators of peripheral myelination. Brain 2017;140:898-913. [Crossref] [PubMed]
- Fledrich R, Stassart RM, Klink A, et al. Soluble neuregulin-1 modulates disease pathogenesis in rodent models of Charcot-Marie-Tooth disease 1A. Nat Med 2014;20:1055-61. [Crossref] [PubMed]
- Fischbach GD. NRG1 and synaptic function in the CNS. Neuron 2007;54:495-7. [Crossref] [PubMed]
- Bartus K, Burnside ER, Galino J, et al. ErbB receptor signaling directly controls oligodendrocyte progenitor cell transformation and spontaneous remyelination after spinal cord injury. Glia 2019;67:1036-46. [Crossref] [PubMed]
- Seshadri S, Faust T, Ishizuka K, et al. Interneuronal DISC1 regulates NRG1-ErbB4 signalling and excitatory-inhibitory synapse formation in the mature cortex. Nat Commun 2015;6:10118. [Crossref] [PubMed]
- Kim N, Min KW, Kang KH, et al. Regulation of retinal axon growth by secreted Vax1 homeodomain protein. Elife 2014;3:e02671. [Crossref] [PubMed]
- Bertuzzi S, Hindges R, Mui SH, et al. The homeodomain protein vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes Dev 1999;13:3092-105. [Crossref] [PubMed]
- Li S, Liu Q, Wang Y, et al. Differential gene expression profiling and biological process analysis in proximal nerve segments after sciatic nerve transection. PLoS One 2013;8:e57000. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Merico D, Isserlin R, Stueker O, et al. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 2010;5:e13984. [Crossref] [PubMed]
- Perisic L, Aldi S, Sun Y, et al. Gene expression signatures, pathways and networks in carotid atherosclerosis. J Intern Med 2016;279:293-308. [Crossref] [PubMed]
- Hou YN, Ding WY, Shen Y, et al. Effect of hyperbaric oxygen on MMP9/2 expression and motor function in rats with spinal cord injury. Int J Clin Exp Med 2015;8:14926-34. [PubMed]
- Deftu AT, Ciorescu R, Gheorghe RO, et al. CXCL1 and CXCL2 Inhibit the Axon Outgrowth in a Time- and Cell-Type-Dependent Manner in Adult Rat Dorsal Root Ganglia Neurons. Neurochem Res 2019;44:2215-29. [Crossref] [PubMed]
- Krishnan A, Purdy K, Chandrasekhar A, et al. A BRCA1-Dependent DNA Damage Response in the Regenerating Adult Peripheral Nerve Milieu. Mol Neurobiol 2018;55:4051-67. [PubMed]
- Wasko NJ, Kulak MH, Paul D, et al. Systemic TLR2 tolerance enhances central nervous system remyelination. J Neuroinflammation 2019;16:158. [Crossref] [PubMed]
- Ko KR, Lee J, Lee D, et al. Hepatocyte Growth Factor (HGF) Promotes Peripheral Nerve Regeneration by Activating Repair Schwann Cells. Sci Rep 2018;8:8316. [Crossref] [PubMed]
- Han D, Chen Y, Kou Y, et al. Profiling of the dynamically alteredgene expression in peripheral nerve injury using NGS RNA sequencing technique. Am J Transl Res 2016;8:871-84. [PubMed]
- Omura T, Omura K, Tedeschi A, et al. Robust Axonal Regeneration Occurs in the Injured CAST/Ei Mouse CNS. Neuron 2015;86:1215-27. [Crossref] [PubMed]
- Mantuano E, Lam MS, Shibayama M, et al. The NMDA receptor functions independently and as an LRP1 co-receptor to promote Schwann cell survival and migration. J Cell Sci 2015;128:3478-88. [Crossref] [PubMed]
- Li Z, Wu F, Xu D, et al. Inhibition of TREM1 reduces inflammation and oxidative stress after spinal cord injury (SCI) associated with HO-1 expressions. Biomed Pharmacother 2019;109:2014-21. [Crossref] [PubMed]
- Kim MS, Shutov LP, Gnanasekaran A, et al. Nerve growth factor (NGF) regulates activity of nuclear factor of activated T-cells (NFAT) in neurons via the phosphatidylinositol 3-kinase (PI3K)-Akt-glycogen synthase kinase 3beta (GSK3beta) pathway. J Biol Chem 2014;289:31349-60. [Crossref] [PubMed]
- Lezana JP, Dagan SY, Robinson A, et al. Axonal PPARgamma promotes neuronal regeneration after injury. Dev Neurobiol 2016;76:688-701. [Crossref] [PubMed]
- Hichor M, Sundaram VK, Eid SA, et al. Liver X Receptor exerts a protective effect against the oxidative stress in the peripheral nerve. Sci Rep 2018;8:2524. [Crossref] [PubMed]
- Pieraut S, Lucas O, Sangari S, et al. An autocrine neuronal interleukin-6 loop mediates chloride accumulation and NKCC1 phosphorylation in axotomized sensory neurons. J Neurosci 2011;31:13516-26. [Crossref] [PubMed]
- Habash T, Saleh A, Roy Chowdhury SK, et al. The proinflammatory cytokine, interleukin-17A, augments mitochondrial function and neurite outgrowth of cultured adult sensory neurons derived from normal and diabetic rats. Exp Neurol 2015;273:177-89. [Crossref] [PubMed]
- Parrinello S, Napoli I, Ribeiro S, et al. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell 2010;143:145-55. [Crossref] [PubMed]
- Kisiswa L, Osorio C, Erice C, et al. TNFalpha reverse signaling promotes sympathetic axon growth and target innervation. Nat Neurosci 2013;16:865-73. [Crossref] [PubMed]
- Xu C, Funahashi Y, Watanabe T, et al. Radial Glial Cell–Neuron Interaction Directs Axon Formation at the Opposite Side of the Neuron from the Contact Site. The Journal of Neuroscience 2015;35:14517-32. [Crossref] [PubMed]
- Duraikannu A, Krishnan A, Chandrasekhar A, et al. Beyond Trophic Factors: Exploiting the Intrinsic Regenerative Properties of Adult Neurons. Front Cell Neurosci 2019;13:128. [Crossref] [PubMed]
- Bota O, Fodor L. The influence of drugs on peripheral nerve regeneration. Drug Metab Rev 2019;51:266-92. [Crossref] [PubMed]
- Tsarouchas TM, Wehner D, Cavone L, et al. Dynamic control of proinflammatory cytokines IL-1β and TNF-α by macrophages in zebrafish spinal cord regeneration. Nature Communications 2018;9:4670. [Crossref] [PubMed]
- Sarhane KA, Ibrahim Z, Martin R, et al. Macroporous nanofiber wraps promote axonal regeneration and functional recovery in nerve repair by limiting fibrosis. Acta Biomater 2019;88:332-45. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


