
Small nucleolar RNA host gene 22 (SNHG22) promotes the progression of esophageal squamous cell carcinoma by miR-429/SESN3 axis
Introduction
It has been studied that esophageal squamous cell carcinoma (ESCC) is one of the main subtypes of esophageal cancer, and ESCC has been proved as cancer due to its high metastasis ability, which caused a significantly high death rate (1). Even though achievements have been made in the treatment of ESCC, the five-year survival rate of ESCC patients was still less than 9%. Meanwhile, the terminal stages of ESCC patients have a poor prognosis (2-4). As early stage ESCC patients have no apparent symptoms, ESCC is diagnosed until an advanced stage with ESCC cell migration, invasion, and metastasis (3,5). Therefore, many scholars have studied the molecular mechanism of ESCC. MicroRNA-134 can prevent the development of ESCC through the MAPK signal pathway mediated by PLXNA1 (6). Overexpression of LEF1 promotes the tumorigenicity of ESCC by activating TGF-β signaling pathway (7). S1PR1 can enhance the proliferation and inhibit the apoptosis of ESCC cells by activating STAT3 signaling pathway (8).
Long noncoding RNAs (lncRNAs) have been reported as a novel RNA that has over 200 nucleotides and lacking the ability of coding proteins (9). Many studies have indicated that lncRNAs are vitally crucial in modulating the biological progress of cancer, including cell proliferation, apoptosis, and autophagy (10-13). lncRNA’s aberrant expression is connected to various cancers, including gastric cancer and ESCC (14,15). For example, HOTTIP inhibition could reduce the growth and invasion of ESCC cells (16). CASC9 was up-regulated in ESCC cells, and silenced CASC9 decreased the migration of ESCC cells (17). LncRNA MEG3 reduces cell growth and EMT by p53 signaling pathway in gastric cancer (18). Although many lncRNAs have been investigated in ESCC progression, the full image of the lncRNA’s role in ESCC should be investigated more. LncRNA small nucleolar RNA host gene 22 (SNHG22) has been studied playing the oncogene role in ovarian epithelial carcinoma (19). At present, there are many related studies on ESCC. However, we still know a few about the function of SNHG22 in ESCC. In this study, the inner mechanism and function of SNHG22 in ESCC will be thoroughly researched.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5332).
Methods
Cell culture and transfection
Human ESCC cells (Kyse-150, TE-10, Eca109) and human esophageal epithelial cell (HET-1A), from ATCC (Rockville, Maryland, MD, USA) are grown at 37 °C with 5% CO2. RPMI 1640 medium (Gibco, Grand Island, NY, USA) was used for cell culture with 10% FBS (Gibco) and 1% antibiotics. The medium was changed every three days.
Quantitative real-time PCR (qRT-PCR)
The total RNAs were acquired from TE-10 and Eca109 cell samples using the Trizol standard protocol for the cDNA synthesis. A qPCR quantified RNA’s expression with SYBR Green PCR Master Mix (Invitrogen, Carlsbad, CA, USA) on the Step-One Plus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The comparative 2−∆∆Ct method was applied for calculating gene expression, with U6 and GAPDH as internal controls.
Cell counting kit-8 (CCK-8)
TE-10 and Eca109 cell samples were prepared in the 96-well plates for CCK-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) as instructed by the supplier. Cell viability was gained using a microplate reader to detect the absorbance at 450 nm.
Colony formation
Cell samples of TE-10 and Eca109 in the 6-well plates were incubated for 2 weeks at 37 °C with 5% CO2. After fixing in 4% formaldehyde and dying in 0.1% crystal violet, clones were counted.
Flow cytometer analysis
ESCC cell samples were reaped and fixed with 4% formaldehyde on ice for 1 h, then mixed with RNase for 30 min. For cycle analysis, samples were cultured with Propidium iodide (PI), then subjected to a flow cytometer using FACSCalibur (BD Biosciences, San Jose, CA, USA). The percentages of a cell sample in G0/G1, S, and G2/M phases were measured. For apoptosis analysis, samples were double-stained for 15 min with Annexin V-labeled with 7AAD and PE (BD Biosciences), finally exposed to flow cytometer.
Immunofluorescence (IF)
ESCC cell samples on the culture slides were first rinsed thrice in phosphate-buffered saline (PBS), then fixed and blocked in 5% BSA in PBS. The primary antibody against LC3B and the secondary antibody is used in sequence. Followed by DAPI staining for the detection of cell nuclei, samples were exposed to the confocal imaging system (Olympus, Tokyo, Japan).
Western blot
The cellular protein extracts from ECSS cell samples were acquired and treated with 12% SDS-polyacrylamide gel electrophoresis (PAGE) gel, shifted to PVDF membranes. After the blockade, samples were probed with primary antibodies against the loading control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and LC3B-I, LC3B-II, P62, and their appropriate HRP-tagged secondary antibodies (all, Abcam, Cambridge, MA, USA). After washing thrice in TBST, western bands were exposed to the electrochemiluminescence (ECL) system (Amersham Pharmacia, Piscataway, New Jersey).
FISH
The fixed cell samples were rinsed in PBS for culturing with 40 nM of SNHG22 FISH probe (Ribobio, Shanghai, China) in a hybridization buffer. After DAPI dyeing, a confocal imaging system analyzed slides.
Subcellular fractionation
Cell samples were washed in PBS, then cultured in a cell fractionation buffer and cell disruption buffer. SNHG22 level in the isolated nuclear and cytoplasmic fractions is determined via qRT-PCR.
RNA immunoprecipitation (RIP)
The RIP assay was achieved by using Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) as guided by the supplier. The antibodies specific to human Ago2 and normal control IgG were acquired from Millipore. Precipitates finally collected by beads were assayed by qRT-PCR.
Dual-luciferase reporter assays
The wild-type (WT) and mutated (Mut) miR-429 binding sites contained in SNHG22 or SESN3 fragments were acquired and inserted into pmirGLO vectors (Promega, Madison, WI, USA). The recombinant plasmids SNHG22-WT/Mut and SESN3-WT/Mut were established and co-transfected with miR-429 mimics or NC-mimics for 48 h. Dual-Luciferase Reporter Assay System (Promega) was finally used for the analysis of luciferase activity.
In vivo tumor growth assay
We transfected 5×106 ESCC cell samples, injected into the 6-week-old male BALB/c-nude mice (Chinese Academy of Medical Science, Beijing, China), with tumor volume measured every four days. A 28-day post-intraperitoneal injection, tumors were dissected from the sacrificed mice for weighing. In vivo tumor growth assay was implemented in accordance with the guidelines for animal care and approved by the Committee on the Ethics of Animal Experiments of Affiliated Hospital of Zunyi Medical University [(2018)1-057].
Statistical analysis
Results from the independent bio-triplications were all given as the mean ± SD. Group difference was processed through a two-tailed Student’s t-test and one-way ANOVA by the application of Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Statistical significance was specified as the P value of <0.05.
Results
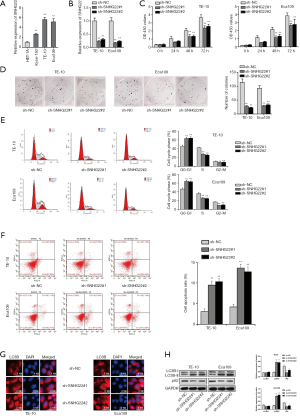
Inhibited SNHG22 could suppress the progression of ESCC cells
SNHG22 has been studied playing the oncogene role in ovarian epithelial carcinoma (16). In this study, we will search for the role of SNHG22 in ESCC. Firstly, the qRT-PCR assay was implemented for assessing the expression of SNHG22 in human esophageal epithelial cells and ESCC cells (Figure 1A). Then, we found that SNHG22 was highly expressed in ESCC cells (Kyse-150, TE-10, and Eca109), especially in TE-10 and Eca109. Thence, TE-10, and Eca109 cells were chosen for the following experiments. For detecting the biological behavior of TE-10 and Eca109 cells when SNHG22 was inhibited, we conducted a qRT-PCR assay to search the inhibition efficiency of SNHG22 (Figure 1B). Then, CCK8 and colony formation assay was implemented to evaluate the proliferation of TE-10 and Eca109 cells when SNHG22 was silenced (Figure 1C,D). We found that OD 450 values and numbers of colonies were both suppressed by SNHG22 inhibition, which indicated that knockdown of SNHG22 could reduce the cell growth of TE-10 and Eca109 cells. Also, flow cytometry analysis was conducted to test the cell cycle progress and apoptosis of TE-10 and Eca109 cells when SNHG22 was inhibited (Figure 1E,F). Results indicated that G0-G1 progress was increased. However, S and G2-M were decreased when SNHG22 inhibition was transfected in. and cell apoptosis rate of TE-10 and Eca109 cells increased much among silenced SNHG22. Thence, inhibited SNHG22 could suppress the cell cycle progress and elevate the apoptosis of TE-10 and Eca109 cells. It has been well proved that LC3B and P62 could act as autophagy markers in cells (20). Furthermore, IF assay and western blot assay were conducted to detect the autophagy of TE-10 and Eca109 cells (Figure 1G,H). LC3B expression and protein level were increased with inhibited SNHG22, and the P62 protein level was reduced. In conclusion, inhibited SNHG22 could suppress the proliferation and cell cycle progress of ESCC cells. Meanwhile, inhibited SNHG22 could increase the apoptosis and autophagy of ESCC cells.
MiR-429 could bind to SNHG22 and act as a tumor suppresser in ESCC cells
It has been proved that SNHG22 affects the chemoresistance of ovarian epithelial carcinoma by sponging miR-2467 (19). Thence, we searched the location of SNHG22 in TE-10 and Eca109 cells by FISH assay and subcellular fraction assay (Figure 2A,B). We found that SNHG22 was found in the cytoplasm of TE-10 and Eca109 cells. Thence we predict lncRNA can act as competing endogenous RNAs to regulate the expression of miRNA and mRNA, which forms as a pathway of lncRNA/miRNA/mRNA (21). Furthermore, we searched the ENCORI website and detected miRNAs that could bind to SNHG22.
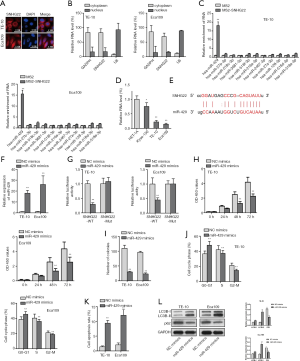
Meanwhile, an MS2-RIP assay was conducted to know about the most potential miRNA (Figure 2C). We found that miR-429 was mostly enriched in MS2-SNHG22. Next, we assessed the expression of miR-429 in TE-10 and Eca109 cells by qRT-PCR assay (Figure 2D). Also, the binding site of SNHG22 and miR-429 was presented according to the ENCORI website (Figure 2E). After we assessed the overexpression efficiency of miR-429 by qRT-PCR assay (Figure 2F), we implemented a luciferase reporter assay to assess the binding relationship in SNHG22 and miR-429 (Figure 2G). Then, CCK8 and colony formation assay illustrated the proliferation of TE-10 and Eca109 cells when miR-429 was overexpressed (Figure 2H,I). We found that overexpressed miR-429 decreased the cell proliferation of TE-10 and Eca109 cells. Next, flow cytometry analysis was used to delineate the cell cycle progress and apoptosis of TE-10 and Eca109 cells (Figure 2J,K). Also, results showed that the gain of function of miR-429 suppressed the cell cycle progress and enhanced the apoptosis of TE-10 and Eca109 cells.
Furthermore, the western blot assay was conducted to verify the autophagy of TE-10 and Eca109 cells when miR-429 was overexpressed (Figure 2L). We found that LC3B-II levels were elevated, and P62 protein levels were decreased by overexpressed miR-429. In conclusion, miR-429 could bind to SNHG22 and act as a tumor suppresser in ESCC cells.
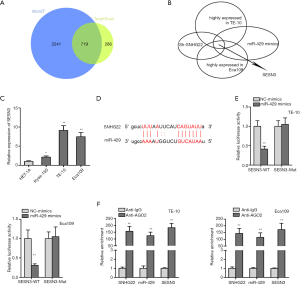
SNHG22/miR-429 /SESN3 axis was proved in ESCC cells
Therefore, we searched the mRNAs that are binding to miR-429 (Figure 3A). Seven hundred nineteen mRNAs were found out. Furthermore, for testing which mRNA was highly expressed in TE-10 and Eca109 cells and could be affected by SNHG22 and miR-429, we conducted qRT-PCR assay (Figure 3B); then, SESN3 was found out. Meanwhile, SESN3 expression was evaluated in the human esophageal epithelial cell (HET-1A) and ESCC cells (Kyse-150, TE-10, and Eca109) via the qRT-PCR assay (Figure 3C). We found that SESN3 was highly expressed in TE-10 and Eca109 cells compared to normal human esophageal epithelial cells. Thence, the binding site of SESN3 and miR-429 was presented according to ENCORI (Figure 3D). Then, luciferase reporter assay and RIP assay found the binding of SESN3 and miR-429, SNHG22, and miR-429. SESN3 were both enriched in Anti-AGO2 of TE-10 and Eca109 cells (Figure 3E,F). In conclusion, SNHG22/miR-429 /SESN3 axis was proved in ESCC cells.
SNHG22/miR-429/SESN3 axis modulated the progression of ESCC cells
Next, we probed the function of the SNHG22/miR-429/SESN3 axis. We firstly detected the inhibition efficiency of miR-429 and the overexpression efficiency of SESN3 (Figure 4A,B). Also, we assessed the expression of SESN3 when SNHG22 is silenced, or miR-429 is overexpressed (Figure 4C). Subsequently, we found that SESN3 expression was suppressed by silenced SNHG22 and overexpressed miR-429. Subsequently, rescue assays were implemented. CCK8 and colony formation assay was implemented to detect the proliferation (Figure 4D,E). Results showed that inhibited cell proliferation of TE-10 and Eca109 cells affected by SNHG22 inhibition was offset by miR-429 inhibition and SESN3 overexpression.
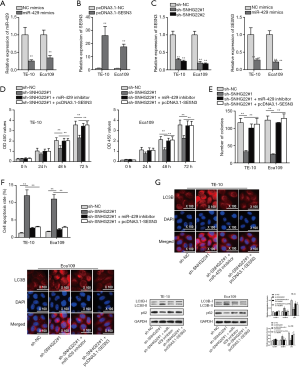
Meanwhile, flow cytometry analysis found that increased cell apoptosis caused by silenced SNHG22 was countervailed by miR-429 knockdown and SESN3 overexpression (Figure 4F). Also, IF assay and western blot assay found that up-regulated LC3B expression, LC3BII protein level, and decreased p62 protein concentration by knockdown of SNHG22 were offset by miR-429 knockdown and SESN3 overexpression (Figure 4G,H). Thence, we concluded that the SNHG22/miR-429/SESN3 axis modulated the proliferation, apoptosis, and autophagy of ESCC cells.
Inhibited SNHG22 could suppress the progression of ESCC in vivo
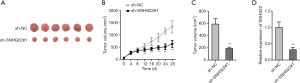
We further implemented vivo experiments for testing the influence of SNHG22 on ESCC progression. The photos of the ESCC tumor were presented when SNHG22 was silenced (Figure 5A). We found that the tumor size in the inhibited SNHG22 group was much smaller than the normal tumor group. Then, tumor volume and weight were both decreased by inhibiting SNHG22 (Figure 5B,C). Meanwhile, the expression of SNHG22 was significantly decreased in inhibited SNHG22 group (Figure 5D). In a word, inhibiting SNHG22 could suppress the progression of ESCC in vivo.
Discussion
Many studies have indicated that lncRNAs expression is dysregulated in various cancers, and they are contributing different functions in modulating the pathological progression of cancers, including cell growth, apoptosis, migration, and metastasis (22,23). Take ESCC as an example; lncRNA SNHG16 enhances cell growth via Wnt signaling pathway way and represents unfavorable prognosis of ESCC (24). Meanwhile, LncRNA GHET1 could activate the EMT progression of ESCC cells to elevate the cell growth and migration in ESCC (25). Also, lncRNA CASC9 could enhance the progression of ESCC cells by modulating the expression of PDCD4 (26). LncRNA SNHG22 has been studied playing the oncogene role in ovarian epithelial carcinoma (19). In this study, we carefully researched the role of SNHG22 in ESCC cells. First, the qRT-PCR assay was implemented to detect the expression of SNHG22 in ESCC cells. We found that SNHG22 was highly expressed. Thence, after detecting the inhibition efficiency of SNHG22, we conducted a series of functional assays to find out the impact of SNHG22 inhibition on the biological function of ESCC cells. CCK8 and colony formation assay found that the proliferation of ESCC cells was decreased by inhibiting SNHG22. Further flow cytometry analysis showed that silenced SNHG22 inhibited the cell cycle progression and increased the apoptosis of ESCC cells.
Meanwhile, IF assay and western blot detected the autophagy of ESCC cells. Further, we found that SNHG22 deficiency suppressed the autophagy of ESCC cells. In a word, SNHG22 could promote cell proliferation, cell cycle progress, and autophagy and decrease cell apoptosis in ESCC cells.
It has been studied that lncRNA can act as competing endogenous RNAs (ceRNAs) to regulate the expression of miRNA and mRNA, which forms as a pathway of lncRNA/miRNA/mRNA (21). Also, studies indicating the lncRNA/miRNA/mRNA pathway could play the function of a regulator of cancer cell biological behavior (27). For example, LncRNA CASC9 enhances the metastasis of ESCC via regulating the expression of LAMC2 (28). LncRNA-TUSC7 modulates the chemoresistance of ESCC cells by sponging miR-224 (29). LncRNA neat1 regulates the viability and invasion of ESCC cells via miR-129/CTBP2 axis (30). LncRAN MNX1-AS1 promotes the development of esophageal squamous cell carcinoma by regulating miR-34a/SIRT1 axis (31). MiR-429 inhibits ESCC proliferation through RAB23/NF-κB pathway and migration mediated EMT (32). Also, SNHG22 affects the chemoresistance of ovarian epithelial carcinoma by sponging miR-2467 (19). In this study, we found the pathway of SNHG22/miR-429/SESN3 in ESCC cells. RIP assay and luciferase reporter assay found that miR-429 could bind to SNHG22 and SESN3. Meanwhile, functional assays detected that overexpressed miR-429 suppresses the progression of ESCC cells either. More importantly, a rescue assay detected that the SNHG22/miR-429/SESN3 axis could modulate the progression of ESCC cells. Also, vivo experiments were conducted, and SNHG22 and miR-429 were proved as oncogene and tumor suppresser in ESCC. In a word, SNHG22 promotes the progression, cell cycle progress, and autophagy of ESCC by the miR-429/SESN3 axis.
Acknowledgments
Thank you very much for the technical support of Guangzhou Yujia Biological Technology Co., Ltd.
Funding: This work was supported by National Natural Science Foundation of China Grants (no. 81860441) and Guizhou Province Science and Technology Cooperation Grants {no. LH[2015]7486}.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5332
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5332
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5332). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were performed in accordance with the guidelines for animal care and was approved by Committee on the Ethics of Animal Experiments of Affiliated Hospital of Zunyi Medical University [(2018)1-057].
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med 2014;371:2499-509. [Crossref] [PubMed]
- van der Wilk BJ, Eyck BM, Lagarde SM, et al. The optimal neoadjuvant treatment of locally advanced esophageal cancer. J Thorac Dis 2019;11:S621-31. [Crossref] [PubMed]
- Tu CC, Hsu PK. The frontline of esophageal cancer treatment: questions to be asked and answered. Ann Transl Med 2018;6:83. [Crossref] [PubMed]
- Miyazaki T, Kato H, Fukuchi M, et al. EphA2 overexpression correlates with poor prognosis in esophageal squamous cell carcinoma. Int J Cancer 2003;103:657-63. [Crossref] [PubMed]
- Wang WW, Zhao ZH, Wang L, et al. MicroRNA-134 prevents the progression of esophageal squamous cell carcinoma via the PLXNA1-mediated MAPK signalling pathway. EBioMedicine 2019;46:66-78. [Crossref] [PubMed]
- Zhao Y, Zhu J, Shi B, et al. The transcription factor LEF1 promotes tumorigenicity and activates the TGF-β signaling pathway in esophageal squamous cell carcinoma. J Exp Clin Cancer Res 2019;38:304. [Crossref] [PubMed]
- Liu Y, Zhi Y, Song H, et al. S1PR1 promotes proliferation and inhibits apoptosis of esophageal squamous cell carcinoma through activating STAT3 pathway. J Exp Clin Cancer Res 2019;38:369. [Crossref] [PubMed]
- Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol 2013;10:925-33. [Crossref] [PubMed]
- Liu B, Li J, Liu X, et al. Long non-coding RNA HOXA11-AS promotes the proliferation HCC cells by epigenetically silencing DUSP5. Oncotarget 2017;8:109509-21. [Crossref] [PubMed]
- Jin L, He Y, Tang S, et al. LncRNA GHET1 predicts poor prognosis in hepatocellular carcinoma and promotes cell proliferation by silencing KLF2. J Cell Physiol 2018;233:4726-34. [Crossref] [PubMed]
- Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet 2010;19:R152-61. [Crossref] [PubMed]
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 2011;10:38. [Crossref] [PubMed]
- Li X, Wu Z, Mei Q, et al. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer 2013;109:2266-78. [Crossref] [PubMed]
- Hao Y, Wu W, Shi F, et al. Prediction of long noncoding RNA functions with co-expression network in esophageal squamous cell carcinoma. BMC Cancer 2015;15:168. [Crossref] [PubMed]
- Chen X, Han H, Li Y, et al. Upregulation of long noncoding RNA HOTTIP promotes metastasis of esophageal squamous cell carcinoma via induction of EMT. Oncotarget 2016;7:84480-5. [Crossref] [PubMed]
- Pan Z, Mao W, Bao Y, et al. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med 2016;5:2442-7. [Crossref] [PubMed]
- Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci 2017;21:3850-6. [PubMed]
- Zhang PF, Wu J, Luo JH, et al. SNHG22 overexpression indicates poor prognosis and induces chemotherapy resistance via the miR-2467/Gal-1 signaling pathway in epithelial ovarian carcinoma. Aging (Albany NY) 2019;11:8204-16. [Crossref] [PubMed]
- Adams O, Dislich B, Berezowska S, et al. Prognostic relevance of autophagy markers LC3B and p62 in esophageal adenocarcinomas. Oncotarget 2016;7:39241-55. [Crossref] [PubMed]
- Xia T, Liao Q, Jiang X, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep 2014;4:6088. [Crossref] [PubMed]
- Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer 2016;139:269-80. [Crossref] [PubMed]
- Yuan SX, Yang F, Yang Y, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology 2012;56:2231-41. [Crossref] [PubMed]
- Han GH, Lu KJ, Wang P, et al. LncRNA SNHG16 predicts poor prognosis in ESCC and promotes cell proliferation and invasion by regulating Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci 2018;22:3795-803. [PubMed]
- Liu H, Zhen Q, Fan Y. LncRNA GHET1 promotes esophageal squamous cell carcinoma cells proliferation and invasion via induction of EMT. Int J Biol Markers 2017;32:e403-8. [Crossref] [PubMed]
- Wu Y, Hu L, Liang Y, et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer 2017;16:150. [Crossref] [PubMed]
- Ergun S, Oztuzcu S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumour Biol 2015;36:3129-36. [Crossref] [PubMed]
- Liang Y, Chen X, Wu Y, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ 2018;25:1980-95. [Crossref] [PubMed]
- Chang ZW, Jia YX, Zhang WJ, et al. LncRNA-TUSC7/miR-224 affected chemotherapy resistance of esophageal squamous cell carcinoma by competitively regulating DESC1. J Exp Clin Cancer Res 2018;37:56. [Crossref] [PubMed]
- Li Y, Chen D, Gao X, Li X, Shi G. LncRNA NEAT1 Regulates Cell Viability and Invasion in Esophageal Squamous Cell Carcinoma through the miR-129/CTBP2 Axis. Dis Markers 2017;2017:5314649.
- Chu J, Li H, Xing Y, et al. LncRNA MNX1-AS1 promotes progression of esophageal squamous cell carcinoma by regulating miR-34a/SIRT1 axis. Biomed Pharmacother 2019;116:109029. [Crossref] [PubMed]
- Wang Y, Yu XJ, Zhou W, Chu YX. MicroRNA-429 inhibits the proliferation and migration of esophageal squamous cell carcinoma cells by targeting RAB23 through the NF-κB pathway. Eur Rev Med Pharmacol Sci 2020;24:1202-10. [PubMed]
(English Language Editor: J. Chapnick)


