
CircRNA-0008717 promotes cell proliferation, migration, and invasion by regulating miR-203/Slug in esophageal cancer cells
Introduction
Esophageal cancer (EC) is one of the most common causes of cancer-related deaths worldwide (1). Although the treatment of EC has been dramatically improved in recent years, the prognosis of EC remains unsatisfactory (2). Radiation therapy (RT) is a vital part of multimodal EC therapy (3), and according to several meta-analyses, patients treated with neoadjuvant chemotherapy or combined chemoradiotherapy before EC surgery have a higher rate of survival than those treated with surgery alone(4). However, the response to these treatments varies from individual to individual, and the treatment for patients with inadequate response to chemotherapy may be ineffective (5,6). Furthermore, radiotherapy resistance often leads to subsequent recurrence and metastasis (5,6). Therefore, it is vital to set up the mechanisms that underlie the progression of EC and develop improved therapeutic strategies.
Circular RNAs (circRNAs) form a covalently closed loop structure and constitute a new class of conserved, stable non-coding RNAs (ncRNAs) that are widely expressed in eukaryotes (7,8). Recently, considerable efforts have been made to understand the role of circRNAs in tumor development (9). Many circRNAs can act as sponges that inhibit the activity of small ncRNAs, microRNAs (miRNAs), which are involved in the post-transcriptional regulation of gene expression (10). CircRNAs have been linked to the progression of multiple diseases, including gastric cancer (11), breast cancer (12), colorectal cancer (13), and malignant melanoma (2). CircRNA-0008717 dysregulation has been implicated in several types of cancer; however, its role in EC progression still is unclear.
Gene expression can be modulated through complementary base-pairing between miRNAs and the 3'-untranslated regions (UTRs) of their target mRNAs, which destabilizes mRNA and prevents translation (14). MiRNAs have increasingly been linked to processes involved in tumorigenesis, including cell growth, apoptosis, migration, and carcinogenesis (15). Therefore, characterizing the role of the large miRNA family is essential for cancer studies (16). Large numbers of miRNAs have been shown as tumor suppressors. In particular, miR-203 has been implicated in several human cancers by targeting the Slug transcription factor (17).
Epithelial-mesenchymal transition (EMT) is a necessary process in tumor development that is regulated by multiple transcription factors, including the Snail family (18). This Slug is known to promote the suppression of E-cadherin, which acts as a crucial tumor suppressor by regulating EMT. The loss of E-cadherin results in dysfunctional cell-to-cell adhesion leading to increased cancer invasion (19,20). In addition, Slug has been reported to involve in a diverse number of processes ranging from tumor cell invasion and metastasis to cell survival and proliferation (21). miR-429 suppresses the cell migration and invasion by targeting Slug in esophageal squamous cell carcinoma (22). Slug inhibition may represent a novel strategy for treatment of esophageal adenocarcinoma (23). However, the effects of miR-203 on cell migration and invasion by targeting Slug in EC remain unclear.
In this study, we investigated the role of circRNA-0008717 in EC in vitro. We found evidence that circRNA-0008717 decreased Slug expression by sponging the miR-203, which promoted the proliferation, invasion, and migration of EC cells. Therefore, circRNA-0008717 could be a suitable therapeutic target to improve the prognosis of EC.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5205).
Methods
Cell cultures
Human normal esophageal epithelial cell line (Het-1A) and human EC cell lines (EC109 and KYSE-150) were obtained from the American Type Culture Collection (ATCC, Manassas, USA). All cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA) with 10% FBS and 1% streptomycin/penicillin in a 5% CO2 incubator at 37 °C.
Cell transfection and grouping
EC109 and KYSE-150 cells at a density of 1×105 cells/well were seeded in 6-well plates. After 24 h of cell adherence, the medium was removed. The miR-203 mimics and scrambles negative control RNA (NC-mimics), miR-203 inhibitor and scrambled negative control RNA (NC-inhibitor), the siRNA for circRNA-0008717 (targeted covalent closed junction), Slug were chemically synthesized by GenePharma (GenePharma, Shanghai, China). The si-circRNA-0008717 sequence was as follows: 5'-TAGAAGACCATGGGGGATGTCAAGAGCATCCCCCATGGTCTTCTATTTTTT-3'. si-Slug sequence was as follows: 5'-TACATGGAGATGTCGAGCACCAT-3'. The full-length sequences of Slug were respectively synthesized and cloned into pcDNA3.1 (GenePharma, Shanghai, China) plasmid to produce pcDNA3.1/Slug (pcDNA Slug). EC109 and KYSE-150 cells were transfected using Lipofectamine® 3000 Reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s protocol. The transfected cells were randomly divided into the si-circRNA group (treated with circRNA-0008717 siRNA), si-NC group (treated with circRNA-0008717 siRNA negative control), miR-203 mimic group (treated with miR-203 mimic), NC-mimic group (treated with miR-203 mimic negative control), miR-203 inhibitor group (treated with miR-203 inhibitor), NC-inhibitor group (treated with miR-203 inhibitor negative control), si-Slug group (treated with Slug siRNA), si-circRNA + miR-203 inhibitor group (treated with circRNA-0008717 siRNA and miR-203 inhibitor), and si-circRNA + Slug group (treated with circRNA-0008717 siRNA and pcDNA Slug). Finally, all cells were incubated at 37 °C for 48 h.
Quantitative real-time PCR (qRT-PCR) assays
Total RNA from EC tissue, normal tissue, or cell lines was extracted by TRIZOL (Invitrogen). A NanoDrop ND-1000 spectrophotometer (NanoDrop, USA) was used to measure the concentration of total RNA. Then, total RNA (500 ng) was reverse-transcribed into cDNA using the PrimeScript RT reagent kit (TaKaRa, Dalian, China) and analyzed by qRT-PCR using an SYBR Green PCR kit (TaKaRa) with the following thermocycling parameters: 95 °C for 3 min and 40 cycles of 95 °C for 15 s followed by 60 °C for 30 s. The following primer sequences are used: circRNA-0008717: Forward: 5'-CTAAGGAGTCACAGGAAGACATC-3'; Reverse: 5'-GTAGAATCTCTCAGACTCAAGGTTG-3'; Slug: Forward: 5'-GCTGTAGGAACCGCCGCCGTGTC-3'; Reverse: 5'-ATTTGTCATTTGGCTTCGGAGTG-3'; GAPDH: Forward: 5'-GAAGGTGAAGGTCGGAGTC-3'; Reverse: 5'-GAAGATGGTGATGGGATTTC-3'. miR-203 (Lot No. 91126N22) and U6 (Lot No. 8300871042) primers were ordered from GenePharma (GenePharma, Shanghai, China). The transcript level of U6 was used to normalize miR-203 expression. GAPDH was used to normalize the transcript levels of circRNA-0008717 and Slug. Relative expression is calculated using the 2-ΔΔCt method (24).
Western blot analysis
Total protein was extracted from EC109 and KYSE-150 cells using RIPA lysis buffer (Sigma, USA). Total protein (50 µg per sample) is separated on a 10% SDS-PAGE gel and transferred onto a polyvinylidene difluoride membrane. Membranes were blocked with 5% non-fat milk for 2 hours and incubated with primary antibodies anti-GAPDH (1:1,000, ab181602, Abcam, UK), anti-Slug (1:1,000, ab51772, Abcam, UK), anti-Vimentin (1:1,000, ab92547, Abcam, UK), or anti-E-cadherin (1:1,000, ab40772, Abcam, UK) at 4 °C overnight. After washing three times, the membranes were incubated with a peroxidase-labeled secondary antibody (anti-rabbit IgG, 1:2,000, ab6721, Abcam, UK) for 2 hours. Enhanced chemiluminescence (ECL) (ThermoFisher, USA) was used to visualize protein bands followed by analysis with Image Lab™ Software (Bio-Rad, USA).
Dual-luciferase reporter gene assay
TargetScan (http://www.targetscan.org/) was used to predict the interaction between circRNA-0008717 and miR-203 and the exact target binding sites. The predicted interaction was examined using a dual-luciferase assay. The wild-type Slug reporter (Slug-Wt) and wild type circRNA-0008717 reporter (circRNA-Wt) were constructed by cloning the 3' UTR of the Slug containing the miR-203 binding site and full-length circRNA-0008717 sequence each into a pGL3 vector (Promega, Madison, WI, USA). GeneArt™ The Site-Directed Mutagenesis System (Thermo Fisher Scientific) was used to generate a mutated circRNA-0008717 reporter (circRNA-0008717-Mut) and a mutated Slug reporter (Slug-mut). Each reporter vector is co-transfected with the miR-203 mimics or miR-203 mimics NC into EC109 and KYSE-150 cells using Lipofectamine 3000. After 48 h, luciferase activity was measured using a dual-luciferase kit (Promega, USA).
Cell counting kit-8(CCK-8) assay
A cell counting kit-8 (CCK-8) kit (Sigma, USA) was used to measure the cell proliferation of EC109 and KYSE-150 cells in 96-well plates (2×104 cells/well). In brief, 10 µL of CCK-8 reagent was added into each well at 24, 48, 72, and 96 hours, and cells were incubated for 1 hour at room temperature. A microplate reader (Bio-Rad, USA) at 450 nm was used to analyze the results.
Transwell assay
Transwell chambers (Corning, USA) were used to detect cell invasion. Briefly, 200 µL of cell suspension (0.1×106 cells) was added to an upper chamber pre-coated with Matrigel (Corning, USA), and the lower chamber contained 600 µL of DMEM with 10% FBS. Cells were incubated for 24 hours at 37 °C. Cells that had migrated to the lower chamber were fixed for 20 minutes in 1% formaldehyde and stained for 20 minutes in crystal violet (0.1%). Stained cells were visualized with a microscope (Olympus), and five randomly selected fields were used to count the number of invading cells.
The scratch wound assay
Transfected EC109 and KYSE-150 cells were seeded into 6-well plates, and a scratch wound assay was used to detect the cell. A wound was introduced to the cell layers using a 200 mL pipette tip, and cells were cultured in 10% FBS-supplemented DMEM. Cell migration was measured at 0 and 48 hours with an inverted microscope.
Cell apoptosis assay
Cell apoptosis was assayed by the flow cytometry (BD Biosciences, USA). After transfection for 24 hours, EC109 and KYSE-150 cells were harvested through trypsinization and then resuspended with PBS buffer. Subsequently, cells were double stained with Annexin V-Alexa Fluor 647 and propidium iodide (PI). Finally, the apoptotic rate was then analyzed using flow cytometer (BD Biosciences, USA).
RNA pull down assay
RNA pull-down were performed as described previously (25). In brief, circRNA-0008717-Wt, circRNA-0008717-Mut and NC were biotinylated to be Bio-circRNA-Wt, Bio-circRNA-Mut and Bio-NC by GenePharma (GenePharma, Shanghai, China). MiR-203-Wt, miR-203-Mut and miR-NC were transcribed using TranscriptAid T7 High Yield Transcription Kit (ThermoFisher Scientific, USA). Biotin RNA labeling mix (GenePharma, China) was used to produce Bio-miR-203-Wt, Bio-miR-203-Mut and Bio-miR-NC. EC109 or KYSE-150 cells were transfected and incubated for 48 hours. After the incubation, cells were lysed with lysis buffer. Then, cell lysate was incubated with Dynabeads M-280 Streptavidin (Invitrogen, CA). RT-qPCR was used to measure purified RNA complex.
Statistical analysis
All data are the mean ± SD from at least 3 independent experiments. A Student’s t-test was used to find significant differences between the two groups, and a one-way ANOVA with Tukey’s post hoc test was used to compare the means of more than two groups. P<0.05 was considered statistically significant. All statistical analyses were performed using SPSS 22.0 (Chicago, IL, USA) and GraphPad Prism 7.0 (GraphPad, San Diego, CA, USA).
Results
The circRNA-0008717 knockdown inhibited the malignant evolution of esophageal cancer cells
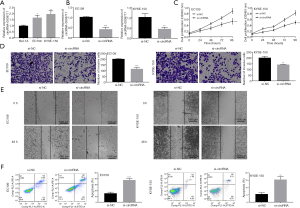
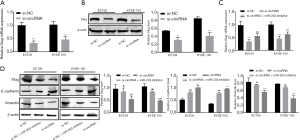
First, we measured the expression of circRNA-0008717 in EC cell lines (EC109 and KYSE-150) to show if circRNA-0008717 participated in EC progression. The qRT-PCR results showed that circRNA-0008717 expression was significantly higher in EC109 and KYSE-150 cells than in the normal cell line Het-1A (P<0.05, P<0.01) (Figure 1A) . These results suggested that circRNA-0008717 was upregulated in EC cells. Second, circRNA-0008717 siRNA (si-circRNA) and its scramble control (si-NC) were separately transfected into EC109 and KYSE-150 cells. As shown by the qRT-PCR results (Figure 1B), the introduction of si-circRNA-0008717 to EC109 and KYSE-150 cells significantly decreased the level of circRNA-0008717 transcripts compared to the si-NC group (P<0.001, P<0.01). Finally, the CCK-8 assay suggested that the circRNA-0008717 knockdown inhibited the proliferation of EC109 and KYSE-150 cells (Figure 1C). Furthermore, the invasion and migration abilities of EC109 and KYSE-150 cells transfected with si-circRNA-0008717 were suppressed (Figure 1D,E). Additionally,as expected, si-circRNA-0008717 significantly enhanced cell apoptosis in both EC109 and KYSE-150 cells (P<0.001, P<0.01, Figure 1F). Therefore, these findings suggest that knocking down circRNA-0008717 inhibited the proliferation, migration, and invasion of EC cells.
CircRNA-0008717 sponged miR-203 in esophageal cancer cells
The expression of miR-203 in EC cell lines was measured by qRT-PCR. The results showed that the level of miR-203 was significantly lower in EC109 and KYSE-150 cells than in Het-1A cells (P<0.001) (Figure 2A). The expression of miR-203 in EC109 and KYSE-150 cells transfected with the miR-203 mimics or miR-203 inhibitor was measured by qRT-PCR. As shown in Figure 2B, the expression of miR-203 in cells in the miR-203 mimic group was significantly higher than in the NC-mimic group (P<0.01). Meanwhile, the expression of miR-203 in cells in the miR-203 inhibitor group was significantly lower than in the NC-inhibitor group (P<0.001). Therefore, the transfection of the miR-203 mimic and miR-203 inhibitor was considered successful. We subsequently used TargetScan to find potential miRNAs that could be targeted by the sponge activity of circRNA-0008717 and found that the seed region of miR-203 contained complementary sequences (Figure 2C). RNA pull-down assay further validated that miR-203 could directly bind with circRNA-0008717 (Figure 2D). The predicted interaction between miR-203 and circRNA-0008717 was confirmed using a luciferase reporter assay. The luciferase activity of the wild type miR-203 significantly inhibited the circRNA-0008717 reporter; however, the activity of the circRNA-0008717 reporter with mutated binding sites was unaffected (Figure 2E).
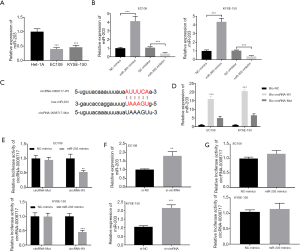
Furthermore, cells transfected with si-circRNA have elevated expression of miR-203 (Figure 2F). However, the luciferase activity of the wild type circRNA-0008717 reporter was unaffected in the miR-203 group (Figure 2G). All these results show that circRNA-0008717 could directly sponge miR-203 in EC cells.
miR-203 targeted Slug in esophageal cancer cells
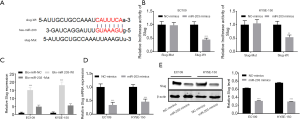
We used Targetscan to find the targets of circRNA-0008717 and miR-203. We found a complementary match between the miR-203 and circRNA-0008717 binding sites and the 3' UTR of Slug (Figure 3A). The interaction between miR-203 and Slug was evaluated using a luciferase reporter assay. In cells transfected with the miR-203 mimic, the luciferase activity of the wild-type Slug vector was significantly lower than the reporter vector with mutated binding sites of Slug (Figure 3B). RNA pull-down assay further validated that miR-203 could directly bind with Slug (Figure 3C). We then examined how miR-203 affected Slug at the transcript and protein levels using qRT-PCR and Western blot. The expression of Slug in EC109 and KYSE-150 cells was inhibited by the miR-203 mimic (Figure 3D,E). These findings suggested that miR-203 directly targeted Slug in EC cells.
CircRNA-0008717 upregulated Slug expression by sponging miR-203
Next sought to set up if sponging of miR-203 by circRNA-0008717 increased the expression of Slug. The qRT-PCR and Western blot results showed that knocking down circRNA-0008717 in EC109 and KYSE-150 cells significantly decreased the levels of Slug mRNA and protein (P<0.001) (Figure 4A,B). Furthermore, co-transfection of the miR-203 inhibitor increased Slug expression in EC cells (Figure 4C). These findings supply evidence for a mechanism by which circRNA-0008717 competitively binds miR-203 and increases the expression of Slug in EC cells. Later, the abundance of two EMT marker proteins was determined by Western blot further to characterize the invasive capabilities of circRNA-0008717 knockdown cells. Knocking down circRNA-0008717 reduced the amount of the mesenchymal marker vimentin and increased the amount of E-cadherin, which it uses as an epithelial marker. However, cells co-transfected with the miR-203 inhibitor had a reverse effect on vimentin and E-cadherin (Figure 4D).
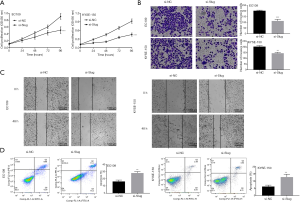
Slug siRNA inhibited the malignant evolution of esophageal cancer cells
To assess the effect of Slug on EC cell phenotype, we performed gain-of-function investigations by transfecting the si-Slug in EC109 and KYSE-150 cells. The CCK-8 assay suggested that si-Slug inhibited the proliferation of EC109 and KYSE-150 cells (P<0.001, Figure 5A). Furthermore, the invasion and migration abilities of EC109 and KYSE-150 were suppressed by si-Slug (P<0.001, P<0.01, Figure 5B,C). Moreover, si-Slug highly enhanced the cell apoptosis of EC109 and KYSE-150 cells (P<0.01, Figure 5D). Therefore, these data reveal that Slug siRNA inhibited the proliferation, migration, and invasion of EC cells.
CircRNA-0008717 promoted the progression of esophageal cancer progression by sponging miR-203 and upregulating Slug
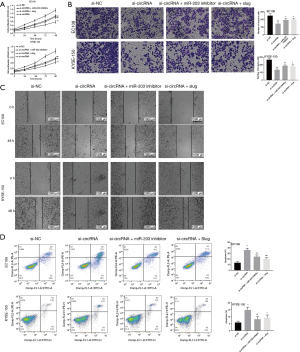
Finally, we investigated if the sponging of miR-203 circRNA-0008717 and increased Slug expression affected the proliferation and invasion of EC cells. We co-transfected the miR-203 inhibitor and circRNA-0008717 siRNA, or co-transfected pcDNA Slug and circRNA-0008717 siRNA into EC109 and KYSE-150 cells to establish if circRNA-0008717 exerted its oncogenic effect through miR-203 to liberate Slug transcripts. The proliferation of EC109 and KYSE-150 was significantly repressed by circRNA-0008717 siRNA. However, the miR-203 inhibitor or pcDNA Slug reduced this effect (Figure 6A). Similarly, the miR-203 inhibitor or pcDNA Slug reduced the migration and invasion of circRNA-0008717 knockdown cells (Figure 6B,C). In addition, the flow cytometry assay showed that the apoptosis of EC109 and KYSE-150 was significantly increased by circRNA-0008717 siRNA. By contrast, the miR-203 inhibitor or pcDNA Slug could rescue it (Figure 6D).
Discussion
The expression of certain circRNAs has been reported in EC tissues and cells by a few previous studies (26,27). However, available information about the specificity and sensitivity of their expression as EC biomarkers, in addition to their functioning networks and mechanisms, is far from sufficient. In this study, we investigated the expression of circRNA-0008717 in two EC cell lines and supplied evidence that circRNA-0008717 sponges miR-203 to liberate Slug transcripts that are targeted by miR-203.
CircRNAs are highly stable and so could prove a useful diagnostic marker or therapeutic target for precision medicine (28,29). A previous study has demonstrated that circRNA-0008717 can promote osteosarcoma progression through sponging miR-203 (29). In our research, we found that the expression of circRNA-0008717 was upregulated and promoted the proliferation, migration, and invasion of EC cells. Consistent with earlier reports, our study supplies evidence that circRNA-0008717 could be a potential therapeutic target to improve the prognosis of EC.
Gene regulation via miRNAs has been implicated in processes that are involved in the development and prognosis of cancers (30). MiRNAs are involved in the post-transcriptional regulation of gene expression by interacting with mRNAs through corresponding miRNA response elements (MRE) (31). In recent years, the sponge role of circRNAs in regulating interactions between miRNAs and oncogenic genes has been realized. First, ciRS-7 was shown to act as a sponge to inhibit the miRNA (miR-7) activity known to have dysregulated expression in cancerous cells (32). Subsequently, it was reported that circRNA-0008717 upregulated Bmi-1 expression by sponging miR-203, which resulted in the progression of osteosarcoma (29). In agreement with this finding, we have shown that circRNA-0008717 sponges miR-203 in EC cells. Previously, the aberrant expression of miR-203 has been linked to multiple malignancies, including EC (33). In line with earlier studies, we found that miR-203 was a key target of circRNA-0008717 and that miR-203 expression was down-regulated in EC.
The Slug transcription factor is a crucial regulator of multiple tumorigenic processes, including cell invasion, metastasis, cell survival, and proliferation (21). In gastric cancer, a reduction in Slug by miRNA-203 was shown to suppress tumor cell proliferation, migration, and invasion (17,34). Here, we also found Slug as a novel target for miR-203 in EC. Furthermore, we found that inhibition of miR-203 increased the expression of Slug and vimentin expression and decreased the expression of E-cadherin. According to previous studies, we know that Slug can increase the invasion of cancer cells in various malignancies by repressing E-cadherin expression (20).
Furthermore, miR-203 inhibits cell motility and invasiveness through Slug/E-cadherin signals (17). In this study, we have shown that circRNA-0008717 can regulate Slug expression via miR-203. Furthermore, transfection with a miR-203 inhibitor reverses the effect of knocking down circRNA-0008717 expression in EC cells. Thus, we have confirmed that circRNA-0008717 can promote the proliferation, migration, and invasion of EC cells by sponging the miR-203.
Our results showed that circRNA-0008717 expression was upregulated in EC cells. Moreover, we have supplied evidence that circRNA-0008717 acts as a sponge that inhibits miR-203, thereby increasing Slug expression, which promotes the proliferation, migration, and invasion of EC cells. This research has proved a novel circRNA regulatory mechanism linked to the progression of EC and has provided a potential therapeutic target that could improve the prognosis of EC patients.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5205
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5205
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5205). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Dicker D, et al. The global burden of cancer 2013. JAMA Oncol 2015;1:505-27. [Crossref] [PubMed]
- Yang YC, Liu GJ, Yuan DF, et al. Influence of exosome-derived miR-21 on chemotherapy resistance of esophageal cancer. Eur Rev Med Pharmacol Sci 2019;23:1513-9. [PubMed]
- Deng W, Lin SH. Advances in radiotherapy for esophageal cancer. Ann Transl Med 2018;6:79. [Crossref] [PubMed]
- Sjoquist KM, Burmeister BH, Smithers BM, et al. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol 2011;12:681-92. [Crossref] [PubMed]
- Maier P, Hartmann L, Wenz F, et al. Cellular pathways in response to ionizing radiation and their targetability for tumor radiosensitization. Int J Mol Sci 2016;17:102. [Crossref] [PubMed]
- Barker HE, Paget JT, Khan AA, et al. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer 2015;15:409-25. [Crossref] [PubMed]
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017;16:94. [Crossref] [PubMed]
- Mitra A, Pfeifer K, Park KS. Circular RNAs and competing endogenous RNA (ceRNA) networks. Transl Cancer Res 2018;7:S624-8. [Crossref] [PubMed]
- Luan W, Shi Y, Zhou Z, et al. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun 2018;502:22-9. [Crossref] [PubMed]
- Nigam S, Manzar N, Ateeq B. Implications of the circular RNAs in localized prostate cancer. Ann Transl Med 2019;7:S195. [Crossref] [PubMed]
- Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015;444:132-6. [Crossref] [PubMed]
- Liang HF, Zhang XZ, Liu BG, et al. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res 2017;7:1566-76. [PubMed]
- Huang G, Zhu H, Shi Y, et al. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One 2015;10:e0131225. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Bueno MJ, Pérez de Castro I, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle 2008;7:3143-8. [Crossref] [PubMed]
- Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene 2006;25:6220-7. [Crossref] [PubMed]
- Gao P, Wang S, Jing F, et al. microRNA-203 suppresses invasion of gastric cancer cells by targeting ERK1/2/Slug/E-cadherin signaling. Cancer Biomark 2017;19:11-20. [Crossref] [PubMed]
- Mittal MK, Singh K, Misra S, et al. SLUG-induced elevation of D1 cyclin in breast cancer cells through the inhibition of its ubiquitination. J Biol Chem 2011;286:469-79. [Crossref] [PubMed]
- Batlle E, Sancho E, Francí C, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2000;2:84-9. [Crossref] [PubMed]
- Uchikado Y, Okumura H, Ishigami S, et al. Increased Slug and decreased E-cadherin expression is related to poor prognosis in patients with gastric cancer. Gastric Cancer 2011;14:41-9. [Crossref] [PubMed]
- Phillips S, Kuperwasser C. SLUG: Critical regulator of epithelial cell identity in breast development and cancer. Cell Adh Migr 2014;8:578-87. [Crossref] [PubMed]
- Zong M, Liu Y, Zhang K, et al. The effects of miR-429 on cell migration and invasion by targeting Slug in esophageal squamous cell carcinoma. Pathol Res Pract 2019;215:152526. [Crossref] [PubMed]
- Zhang K, Zhang S, Jiao X, et al. Slug regulates proliferation and invasiveness of esophageal adenocarcinoma cells in vitro and in vivo. Med Oncol 2011;28:1089-100. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Ji N, Wang Y, Bao G, et al. LncRNA SNHG14 promotes the progression of cervical cancer by regulating miR-206/YWHAZ. Pathol Res Pract 2019;215:668-75. [Crossref] [PubMed]
- Su H, Lin F, Deng X, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med 2016;14:225. [Crossref] [PubMed]
- Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep 2016;6:35576. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Zhou X, Natino D, Qin Z, et al. Identification and functional characterization of circRNA-0008717 as an oncogene in osteosarcoma through sponging miR-203. Oncotarget 2017;9:22288-300. [Crossref] [PubMed]
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014;20:460-9. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Gu J, Wang Y, Wu X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des 2013;19:1292-300. [PubMed]
- Yang L, Liang H, Wang Y, et al. MiRNA-203 suppresses tumor cell proliferation, migration and invasion by targeting Slug in gastric cancer. Protein Cell 2016;7:383-7. [Crossref] [PubMed]


