
MID1 and MID2 regulate cell migration and epithelial-mesenchymal transition via modulating Wnt/β-catenin signaling
Introduction
MID1 gene was firstly identified as a causative gene of the X-linked form of a genetic disorder, Opitz syndrome, characterized by variable facial abnormalities along with other systematic defects. The high incidence of the malformations including cleft lip/palate, hypospadias and defects in cardiac septation indicated that defects in midline fusion and/or tissue remodelling could be the common pathogenesis of these abnormalities (1). For incidence, the proper development of the upper lip and palate depend on the precisely coordinated emigration of neural crest cells and the proliferation of the lateral/medial nasal processes and maxillary processes. The apoptosis and epithelial-mesenchymal transition (EMT) then occur subsequently in the contact zone to ensure the fusion of the facial prominences (2).
Up-to-date, mutations in MID1 have been found in a significant proportion of OS cases (1), including point mutations, deletions and mutations in regulatory regions. However, the variability in clinical phenotype may also suggest the genetic heterogeneity and the existence of other modifying genes. Sequence similarity searches of the EST databases using the hMID1 cDNA revealed a closely related homologue, MID2, which encode a 685-amino-acid protein sharing 76% amino acid identity with MID1. Similar functions have been ascribed to MID1 and MID2 in vitro and in vivo. Both MID1 and MID2 are found distributed along microtubules in all cell types examined and homo- or hetero-dimerization is a prerequisite for their microtubule-association (3,4). Disrupted function of MID1/MID2 resulted in Xenopus neural tube defects due to disorganization of microtubules and functional redundancy of MID1/MID2 has also been displayed in avian left–right determination during early development in chick embryo. Both MID1 and MID2 have E3 ubiquitin ligase activities (5-7). MID1 and MID2 displayed E3 ubiquitin ligase activities with unique and common protein interactors/substrates (6-10). One of the best characterized common MID protein partners is alpha4, which forms a complex with the catalytic subunit of protein phosphatase 2A (PP2Ac) and acts as the regulatory subunit of PP2Ac (4,11). Previous results showed that MID1 downregulated the cellular level of PP2A, which in turn affected the dephosphorylation of MID1 by PP2A (12). However, more recent studies suggested MID1 could directly ubiquitinate PP2A and the presence of alpha4 could actually protect PP2A from degradation by polyubiquitlation (7,13,14). However, evidences were provided that alpha4 could also be poly-/monoubiquitinated by MID1, which in turn switched alpha4’s activity towards PP2A from protective to destructive (7,13,15,16). These results make the cellular functions of MID1 more ambiguous and the molecular etiology of Opitz syndrome more profound.
In this study, MID1/MID2 were overexpressed or down regulated using siRNA in cultured 293T cells and the impact on cellular level of PP2Ac was investigated. It was found that the cellular level of phosphorylated PP2Ac was significantly decreased in cells transfected with siMID1 or siMID2, while the cellular level of total PP2Ac was not affected by either MID1/MID2 silencing or overexpression. Multiple residues of PP2Ac can be phosphorylated and amongst them, phosphorylation of Tyr307 has been well characterized and found to inhibit PP2A’a enzymatic activity (17). As the most abundance phosphatases, PP2A both positively and negatively regulates Wnt/β-catenin signaling via targeting multiple components (18). β-catenin, the main effector of Wnt, can be directly dephosphorylated and stabilized by PP2A (19). The scaffold protein, Axin, of the β-catenin destruction complex, could also be dephosphorylated by PP2A, which in turn affected the assemble of the β-catenin destruction complex and increased the cellular level of β-catenin (20). However, controversial results demonstrated that another component of the β-catenin destruction complex, GSK3β, can also be dephosphorylated by PP2A, resulted in the activation of GSK3β and downregulation of Wnt/β-catenin signaling (21,22).
Given the importance of Wnt in regulating neural crest cell specification and in regulating the fate of epithelium in developing facial prominences (18), the potential influence of MID1/MID2 on Wnt signaling was further investigated in this study. Using Top/Fop flash TCF/LEF luciferase reporter assay, we found that the Wnt signaling was inhibited in cells overexpressing MID1/MID2 and was activated in cells with siRNA mediated knock-down of MID1/MID2. In line with that, the cellular level of β-catenin, the critical effector of Wnt, was downregulated in cells overexpressing MID1/MID2. We further investigated the potential impact of MID1/MID2 on cell migration and elevated cell migration was observed in cells with siRNA mediated knock-down of MID1/MID2 using wound-healing assay.
Collectively, the results presented and discussed in this paper provided some of the first evidence in support of the hypothesis that the Opitz Syndrome gene MID1 and its homologue MID2 have a potential role in regulating Wnt pathway and thus affect epithelium fate and cell migration via its protein interactor PP2Ac. MID1 and MID2 displayed similar cellular function in all the experiments presented in this study, which further support a functional redundancy between these two highly related homologs. The results presented in this study would help to understand the aetiology of the dysregulated craniofacial development seen in Opitz Syndome.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5583).
Methods
Reagents
DMEM and fetal bovine serum (FBS) used in cell culture were purchased from Invitrogen. 3,3-diaminobenzidine tetrachloride (DAB), 1,4-dithiothreitol (DTT), Dimethyl Sulfoxide (DMSO), Triton X-100, Paraformaldehyde, Bovine Serum Albumin, Ethanol, and KCl hematoxylin protease inhibitor cocktail, Penicillin and streptomycin were from Sigma. Blue RangeTM Prestained protein Molecular Marker was from Pierce. BCA Protein Assay Kit was from Thermo Fisher Scientific. Anti-MID1 antibody (cat. no. ab70770; 1:500), anti-MID2 antibody (cat. no. ab14749; 1:500) and anti-β-catenin rabbit polyclonal antibody (cat. no. ab16051; 1:1,000) was purchased from Abcam company. PP2Ac Subunit Rabbit mAb (cat. no. 2259; 1:1,000), β-Actin Mouse mAb (cat. no. 3700; 1:1,000), E-cadherin rabbit mAb (cat. no. #3195, 1:100), Vimentin XP® Rabbit mAb (cat. no. #5741, 1:200) were purchased from Cell Signaling Technology, Inc. Human/Mouse/Rat Phospho-PP2A Catalytic Subunit Antibody (cat. no. AF3989, 1:500) was from Bio-Techne Ltd (Abingdon, UK), HRP-conjugated Goat Anti-Mouse IgG (H+L) (cat. no. SA00001-1; 1:1,000), HRP-conjugated Donkey Anti-Goat IgG (H+L) (cat. no. SA00003-1; 1:3,000) and HRP-conjugated Goat Anti-Rabbit IgG (H+L) (cat. no. SA00001-2; 1:3,000) were purchased from Proteintech (Chicago, Ill., USA). Alexa Fluor® 594 AffiniPure Goat Anti-Rabbit IgG (H+L) (cat. no. 111-585-003, 1:100) were purchased from Jacksons ImmunoResearch (Europe, Ltd. Newmarket, UK). PrimeSTAR DNA Polymerase, EasyTaq DNA Polymerase, EcoRI, XhoI and T4 DNA ligase were purchased from Takara Bio, Inc. (Otsu, Japan).
Plasmids and siRNAs
The entire ORFs of human MID1 and MID2 were amplified with the specific primers (primer sequences as listed in Table 1) from cDNA template prepared from cultured HEK 293T cells. The amplification was performed in 50 µL volumes containing 1 µL of each primer (10 µmol/L), 10 µL 5× PCR mix, 1 µL PremeSTAR® HS DNApolymerase under the following conditions: an initial step of 98 °C for 5 min and then 34 cycles of 98 °C for 30 s, 58 °C for 15 s, 72 °C for 150 s, and a final extension at 72 °C for 5 min. The PCR products were separated by electrophoresis on 1% polyacrylamide gel and the target fragment was purified using an agarose gel extraction kit (Omega). Followed by restriction digestions, the hMID1 and hMID2 ORF were cloned into the XhoI and EcoRI site of pcDNA3.1 or pEGFP-C1 vector, respectively. The plasmids generated were verified by sequencing (Sangon Biotech, Shanghai, China) and stocks were made using EndoFree Maxi Plasmid kit according to the manufacturer’s instructions (Tiangen Biotech Co., Ltd., Beijing, China).

Full table
The sequences of siRNAs used to knock-down the endogenous expression of MID1 and MID2 were listed in Table 1 and the siRNAs were synthesized by GenePharma Co. (Shanghai, China). A non-sense siRNA was used as control in testing the efficiency in MID1 or MID2 gene silencing.
Cell culture and transfection
The HEK293T cells were maintained and cultured in Dulbecco’s modified Eagles medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 µg/mL streptomycin (Biochrom KG, Berlin, Germany) at 37 °C in a humidified 5% CO2 incubator. Transfection of cells with the indicated constructs or siRNA using Lipofectamine™ 2000 transfection reagent (Invitrogen) were conducted according to the manufacturer’s instructions.
Western blotting
Cultured cells were collected and were resuspended in lysis buffer containing 50 mM Tris-HCl (pH7.4), 100 mM KCl, 10% glycerol, 1 mM EDTA, 1% TritonX-100 and 1 mM DTT protease inhibitors (Roche). The supernatants were collected by centrifugation at 12,000 ×g for 15 min at 4 °C. Protein concentration was measured by bicinchoninic protein assay (Life technologies, number 23227). Thirty µg of total cellular proteins of each sample were separated by electrophoresis on 10% SDS-polyacrylamide gels and then transferred to nitrocellulose membranes (Millipore, Billerica, MA). The membrane was blocked with TBST buffer containing 5% skimmed milk powder for 1 h at room temperature before probed with specific primary antibodies at 4 °C overnight. The membrane was washed three times, each for 10 min, with TBST, before incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. The membrane was again washed three times with TBST before monitoring by chemiluminescence (Immobilon horseradish peroxidase; Millipore, Billerica, MA) using Amersham Imager 680 (GE Healthcare Bio-Sciences Corp, Piscataway, NJ). Immunoblotting experiments were performed three times. Densitometry analysis was quantified using ImageJ v1.4.3.67 and the relative expression of the target protein was normalized to internal β-actin.
TCF/LEF luciferase reporter assay
The HEK293T cells were cultured at a density of 2×104 cells/well in 96-well culture plates and were co-transfected with the plasmids pTOP-FLASH (encoding for the luciferase gene preceded by 3 Tcf/Lef binding sites), the constitutively active vector encoding for Renilla luciferase (Promega, Madison, WI, USA) using as co-transfection control, GFP-MID expression vector or siRNA using LipofectamineTM 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. 24 h post-transfection, the reporter activity was assayed using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI) according to the manufacturer’s instruction. The luciferase activity was quantified in a luminometer (SpectraMax M5 spectrophotometer). The ratio of luciferase/renilla activity was obtained and the strength of Wnt/β-catenin signaling was expressed as the relative values with respect to control cells transfected with nonsense siRNA or empty vector. Each sample was prepared in triplicated and data were collected from 3 independent experiments.
Immunofluorescence analysis
The cultured 293T cells were plated on coverslips and transfected with indicated constructs 24 h before analysed. The cells were rinsed twice with PBS and fixed with 4% PFA in PEM buffer (1M PIPES, 20 mM MgCl2, 50 mM EGTA and adjust pH 7.0) for 15 min at room temperature. The cells were subsequently permeabilized with 0.2% Nonidet-P40 in PEM, followed by staining for 1 h at 37 °C with the appropriate dilutions of the indicated primary antibodies (anti-E-cadherin mAb and anti-Vimentin mAb). Excess primary antibodies were washed off with PEM and then incubated with appropriate dilutions of the conjugated secondary antibodies (Alexa Fluor Goat Anti-Rabbit IgG) for 1 h at 37 °C. The coverslips were again washed three time in PEM before mounted on top of 20 µL glycerol containing Hochest 33342 on slides. The slides were visualized under appropriate wavelength light on a Zeiss LSM710 confocal microscope. The images were captured using Zeiss LSM 510 EMTA camera and processed with Zeiss LSM Image Browser 4.0 (Zeiss GmbH, Jena, Germany). A total of nine different views of co-transfected cells were randomly selected and fluorescence intensities were normalized to the un-transfected cells within the same view. Fluorescence quantifications were calculated with ImageJ (National Institutes of Health, Bethesda, MD, USA) and the results were presented as means ± SD.
Wound healing assay
For the wound healing assays, 293T cells were prepared on 6-well plates in biological triplicates. The cells were transfected with indicated plasmids or siRNA at ~60% confluence and incubated overnight to allow to reach 90% confluence for the assay. A wound in each well was made by scratching straight through the middle of the well using a 200-µL pipette tip. Culture media was gently sucked out to remove detached cells and debris. The cells were refed with fresh media and maintained at 37 °C for up to 24 h. The cells were photographed under 100× magnification at 12 h and 24 h post transfection with an inverted fluorescence microscope (Nikon Eclipse TE300; Nikon, Tokyo, Japan). The areas of the cell-free zone into which cells migrated were quantitated under the microscope using ImagePro Plus software (Version 6.0). The migration index was calculated as (area0-areat/area0, in which the underscore “0” refers to time zero and the underscore “t” refers to the indicated time point) and normalized to that of control cells. Experiments were carried out in triplicate and repeated three times.
Statistical analyses
Each experiment was performed three times, independently. All values are expressed as arithmetic means ± SEM (error bars). Statistically significant differences among groups were determined using students t-test. All statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software, San Diego, CA).
Results
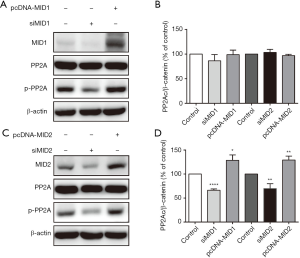
PP2Ac phosphorylation was affected by the change of MID1 or MID2 expression
It was supported by the early laboratory results that MID1 catalyze the ubiquitylation of serine/threonine protein phosphatase 2A (PP2Ac) and drive PP2Ac to ubiquitin-mediated proteasomal degradation via its regulatory subunit α4, which was shown to be a direct protein interactor of MID1 (6). However, recent research revealed a far more complicated interplay of the MID1/α4/PP2Ac complex and its cellular functions. Both PP2Ac and α4 can be ubiquitinated, which in turn, altered α4’s function from protective to disruptive as to PP2Ac degradation (6,7,13). The formation of the MID1/α4/PP2Ac complex also resulted in MID1 dephosphorylation, of which the consequences were not fully understood (12).
Our results presented here that knock-down of the endogenous MID1 expression in cultured 293T cells resulted in significantly reduced phosphorylated PP2Ac, while the change of cellular level of total PP2Ac was not observed. Overexpression of MID1 resulted in unnoticeable change of both total cellular PP2Ac nor phosphorylated PP2Ac (Figure 1A,B). Similar results were observed with knock-down or overexpression of MID2 (Figure 1C,D).
Overexpression of MID1/MID2 downregulated Wnt signalling, possibility via PP2Ac phosphorylation and thereby inactivation
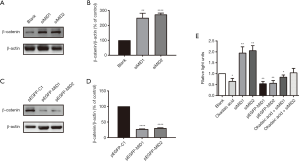
Multiple residues on PP2Ac can be phosphorylated and prevent the subsequent assemble/activation of the holoenzyme. Wnt signaling has been extensively investigated in regulating neural crest cell differentiation and craniofacial development. A/J mice carrying background mutations in Wnt9b displayed high incidence (5–10%) of spontaneous cleft lip and palate, indicating dysregulation of Wnt signaling in the pathogenesis of this malformation (23). Multiple components in Wnt pathway are regulated by phosphorylation and are the potential targets of PP2A. PP2A’s dual regulation, both positive and negative, of Wnt signaling was evidenced by numerous laboratory results. Our results in this study showed that the expression of the β-catenin, the main effector of Wnt, was significantly upregulated in MID1/MID2 knock down cells using siRNA (Figure 2A,B), while reduced in cells overexpressing MID1 or MID2 (Figure 2C,D). We further monitored the potential influences of MID1/MID2 on Wnt signaling in cultured 293T cells using Dual-Luciferase Reporter Assay System (Promega). In line with the results of the above western blot, the Wnt signaling was found to be compromised in cells overexpressing MID1/MID2, while was enhanced in cells with MID1/MID2 gene silencing (Figure 2E). We also noticed that the compromised Wnt signaling in cells treated with Okadaic acid, a specific PP2A inhibitor, and was partially recovered by MID1/MID2 gene silencing, indicating that MID1/MID2 was involved in Wnt signaling via regulating PP2A’s activity.
Induction of EMT was observed in cultured cells with siRNA-mediated knock-down of MID1/MID2
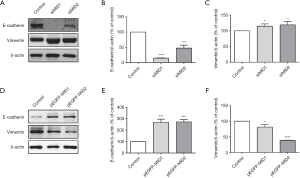
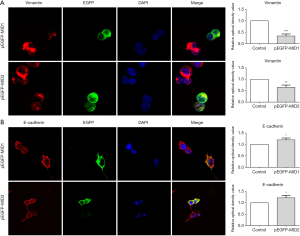
The Wnt/β-catenin signalling pathway shares a key component, β-catenin, with the cadherin-based adhesion system. The cadherin complex is dynamic, allowing for cell-cell rearrangements such as epithelial-mesenchymal transition (EMT), where the complex turns over through internalisation. Cadherins are generally considered negative regulators of Wnt signalling because that a significant amount of β-catenin sequestered by E-Cadherin at the plasma membrane and knock-down of E-Cadherin led to redistribution of β-catenin in cytoplasmic, followed by nuclear translocation and subsequent activation of Wnt signaling. In this study, the expression of the epithelial marker E-cadherin and the mesenchymal marker vimentin were measured by western blot to evaluate the potential influence of MID1/MID2 in regulating EMT. Decreased E-cadherin and increased vimentin were observed in cultured 293T cells with siRNA mediated knock-down of MID1/MID2, and vice versa (Figure 3), reflecting the modulation of EMT. These observations were further confirmed using immunofluorescence, showing increased E-cadherin and decreased vimentin in cells overexpressing GFP-MID1 or GFP-MID2 (Figure 4).
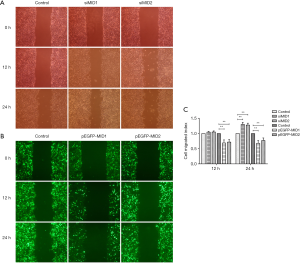
MID1/MID2 gene silencing promoted cell migration
The proper formation of the craniofacial structures relies on the coordinated emigration of neural crest cells, cell proliferation and EMT. During embryonic development Wnt family members and bone morphogenetic proteins (BMPs) cooperatively induce EMT in the neural crest. The process of EMT are usually companied by the conversion of cells with strong cell-cell adhesion to cells with more motile characteristics. Using wound healing assay, our results in this study showed accelerated migration of cells with siRNA-mediated knock-down of MID1/MID2 and decelerated migration of cells overexpressing GFP-MID1 or GFP-MID2 (Figure 5). It suggested that the disturbed cell migration and EMT, as the consequences of defective MID1/MID2 gene function, might be pivotal in the malformation of craniofacial development seen in OS patients.
Discussion
The craniofacial development is a highly organized and orchestrated integration of cell migration, proliferation and EMT in a three-dimensional morphogenesis (24-26). The cranial neural crest cells (CNCCs), originated from the dorsal neural tube, undergo EMT and emigrate from the diencephalon, anterior mesencephalon and rhombomeres, to form the skeletal structures and non-skeletal derivatives, including cartilage, bone, tendons and connective tissue as well as neurons, glia and melanocytes in the head and face (27-29). This morphogenetic process is tightly regulated by the interplay of multiple signaling molecules, which control the fate of cells in both mesenchyme and epithelium of the embryonic facial prominence. Both conventional genetic screening and the more recent high-throughput sequencing revealed a complex of causative genes for craniofacial malformations, as well as altered noncoding RNA expressions (25,30).
MID1 was first identified as the causative gene for X-linked OS, and was found mutated also in non-syndromic cleft lip/palate (31,32). MID1 displayed ubiquitin-E3 ligase activity and early laboratory results supported that MID1 mediated the ubiquitylation and degradation of PP2Ac through direct interaction with alpha4, the regulatory subunit of PP2Ac (6). However, recent research revealed a much more complicate influence of MID1 as to the regulation of PP2A’s activity. Instead of directing PP2A’s ubiquitination and degradation, Alpha4 itself could be ubiquitinated by MID1 and then convert alpha4’s function from protective to disruptive (7,14,15). Besides that, the active transport of MID1 on microtubules can be regulated by the phosphorylation and dephosphorylation of MID1 by PP2A (12). Here, the results in our study showed that MID1 gene silencing significantly reduced phosphorylated PP2Ac and vice versa, although the level of the total cellular PP2Ac was not affected.
PP2Ac phosphorylation results in the inactivation of its enzymatic activity, although the precise phosphorylation sites and their impacts on protein function were not fully understood. Amongst them, the Tyr 307 phosphorylation by a number of receptor and non-receptor tyrosine kinases including the epidermal growth factor receptor (EGFR), insulin receptor (INSR) and protooncogene tyrosine-protein kinase Src (SRC) was well studied and was found to reduce PP2Ac’s activity up to 90% (17,33). Given the fact that multiple protein targets have been found ubiquitylated by MID1, our results indicated the possibility that the protein kinases and phosphatases in regulating PP2Ac’s phosphorylation could also be potential targets of MID1. Recent findings revealed a novel function of MID1 in regulation protein translation via mTOR-eIF signaling (6,34,35). Whether MID1 can regulated the translation of tyrosine kinases and phosphatases and therefore affect the PP2A phosphorylation remain to be clarified with further investigations.
As we mentioned before, proper spatio-temporal development of facial prominence depends on the cross-talk of signaling molecules including Sonic hedgehog, Wnt and BMPs (36). Multiple components of Wnt signaling are subjected to regulation by phosphorylation and therefore, are potential targets of PP2Ac. Both positive and negative regulation of Wnt signaling are supported by numerous evidence. For instance, β-catenin, the key effector of canonical Wnt signaling, can be dephosphorylated by PP2A, lead to stabilization of the protein and the increased Wnt activity (18). In this study, we observed that siRNA-mediated knock-down of MID1 expression significantly increased β-catenin and enhanced Wnt signaling using luciferase reporter assay. It was likely to be the consequence of inhibition of PP2Ac’s phosphorylation and inactivation since the compromised Wnt signaling in cells treated with Okadaic acid, the PP2Ac’s inhibitor, was partially rescued by MID1 gene silencing.
Given that the common key component, β-catenin, is shared by both Wnt and Cadherin pathway, cadherins are generally considered as negative regulators of Wnt signalling by sequestering the cytoplasmic β-catenin on membrane (37). Activation of Wnt signaling is often seen with the downregulated E-Cadherin/β-catenin complex on membrane in both cancer cells and embryonic cells (38,39). Dysregulation of the interplay between these two pathways lead to cell-cell rearrangements and epithelial-mesenchymal transition (EMT), which are fundamental processes in tumour progression as well as in embryonic development (40). In consistent with the increased β-catenin and Wnt signaling, we proved that the E-Cadherin was downregulated and the expression of vimentin, the other marker of EMT, was upregulated in cultured 293T cells with siRNA mediated knock-down of MID1/MID2 expression. Opposite changes were observed in cells overexpressing GFP-MID1 or GFP-MID2. The influences on the expression of molecular markers of EMT were further confirmed using immunofluorescence. In consistent with the promoted EMT, accelerated cell migration was observed with MID1/MID2 gene silencing in cultured 293T cells using wound-healing assay. However, embryonic cell migration is a complex process and needs to be further investigated in vivo. In our unpublished data, we transfected the chick neural crest cells using microinjection of the RCAS retroviral vectors expressing MID1/MID2 siRNA in the neural tube of HH9-10 chick embryo (41). Our preliminary results indicated an elevated emigration of chick neural crest cells with siRNA mediated MID1/MID2 gene silencing, in consistent with the in vitro studies presented in this paper.
Interestingly, a pattern of changes similar to MID1 was noticed for MID2 in all the parallel functional assays in this study. Given the overlapping cellular localization and function presented in previous studies, the results in this research strongly support a functional redundancy of these two highly related homologues. Collectively, we proposed that MID1 and its homolog, MID2, were involved in the regulation of Wnt signaling pathway via modulating PP2A’s phosphorylation/activation. The aetiology of the cranial malformation seen in OS patients with MID1 mutation may attribute to the defective cell migration and EMT, as the consequences of dysregulated Wnt signaling.
Acknowledgments
Special thanks to Dr. Rongjie Yu for kindly providing the pTop/Fop-FLASH plasmids for luciferase assay.
Funding: This work was sponsored by the The Natural Science foundation of Guangdong (2020A1515010160) and the Key Program of Marine biology Special Foundation of Department of Natural Resources of Guangdong Province {GDNRC [2020]035}.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5583
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5583
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5583). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li B, Zhou T, Zou Y. Mid1/Mid2 expression in craniofacial development and a literature review of X-linked Opitz syndrome. Mol Genet Genomic Med 2015;4:95-105. [Crossref] [PubMed]
- Cox TC. Taking it to the max: The genetic and developmental mechanisms coordinating midfacial morphogenesis and dysmorphology. Clin Genet 2004;65:163-76. [Crossref] [PubMed]
- Buchner G, Montini E, Andolfi G, et al. MID2, a homologue of the Opitz syndrome gene MID1: similarities in subcellular localization and differences in expression during development. Hum Mol Genet 1999;8:1397-407. [Crossref] [PubMed]
- Granata A, Savery D, Hazan J, et al. Evidence of functional redundancy between MID proteins: implications for the presentation of Opitz syndrome. Dev Biol 2005;277:417-24. [Crossref] [PubMed]
- Suzuki M, Hara Y, Takagi C, et al. MID1 and MID2 are required for Xenopus neural tube closure through the regulation of microtubule organization. Development 2010;137:2329. [Crossref] [PubMed]
- Trockenbacher A, Suckow V, Foerster J, et al. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat Genet 2001;29:287-94. [Crossref] [PubMed]
- LeNoue-Newton M, Watkins GR, Zou P, et al. The E3 ubiquitin ligase- and protein phosphatase 2A (PP2A)-binding domains of the Alpha4 protein are both required for Alpha4 to inhibit PP2A degradation. J Biol Chem 2011;286:17665-71. [Crossref] [PubMed]
- Han X, Du H, Massiah MA. Detection and characterization of the in vitro e3 ligase activity of the human MID1 protein. J Mol Biol 2011;407:505-20. [Crossref] [PubMed]
- Senese S, Lo YC, Gholkar AA, et al. Microtubins: A novel class of small synthetic microtubule targeting drugs that inhibit cancer cell proliferation. Oncotarget 2017;8:104007-21. [Crossref] [PubMed]
- Zanchetta ME, Meroni G. Emerging Roles of the TRIM E3 Ubiquitin Ligases MID1 and MID2 in Cytokinesis. Front Physiol 2019;10:274. [Crossref] [PubMed]
- Liu D, Schwender H, Wang M, et al. Gene-gene interaction between MSX1 and TP63 in Asian case-parent trios with nonsyndromic cleft lip with or without cleft palate. Birth Defects Research 2018;110:317-24. [Crossref] [PubMed]
- Aranda-Orgillés B, Aigner J, Kunath M, et al. Active transport of the ubiquitin ligase MID1 along the microtubules is regulated by protein phosphatase 2A. PLoS One 2008;3:e3507. [Crossref] [PubMed]
- Du H, Wu K, Alma D, et al. MID1 Catalyzes the Ubiquitination of Protein Phosphatase 2A and Mutations within Its Bbox1 Domain Disrupt Polyubiquitination of Alpha4 but Not of PP2Ac. PLoS One 2014;9:e107428. [Crossref] [PubMed]
- McConnell JL, Watkins GR, Soss SE, et al. Alpha4 Is a Ubiquitin-Binding Protein That Regulates Protein Serine/Threonine Phosphatase 2A Ubiquitination. Biochemistry 2010;49:1713-8. [Crossref] [PubMed]
- Du H, Huang Y, Zaghlula M, et al. The MID1 E3 ligase catalyzes the polyubiquitination of Alpha4 (α4), a regulatory subunit of protein phosphatase 2A (PP2A): novel insights into MID1-mediated regulation of PP2A. J Biol Chem 2013;288:21341-50. [Crossref] [PubMed]
- Latta EJ, Golding JP. Regulation of PP2A activity by Mid1 controls cranial neural crest speed and gangliogenesis. Mech Dev 2012;128:560-76. [Crossref] [PubMed]
- Farrington CC, Yuan E, Mazhar S, et al. Protein phosphatase 2A activation as a therapeutic strategy for managing MYC-driven cancers. J Biol Chem 2020;295:757. [Crossref] [PubMed]
- Thompson MD, Moghe A, Cornuet P, et al. β-catenin regulation of farnesoid X receptor signaling and bile acid metabolism during murine cholestasis. Hepatology 2018;67:955-71. [Crossref] [PubMed]
- Zhang W, Chen X, Yu T, et al. PR55 alpha, a regulatory subunit of PP2A, specifically regulates PP2A-mediated beta-catenin dephosphorylation. J Biol Chem 2009;284:22649. [Crossref] [PubMed]
- Strovel ET, Wu DQ, Sussman DJ. Protein Phosphatase 2Cα Dephosphorylates Axin and Activates LEF-1-dependent Transcription. J Biol Chem 2000;275:2399-403. [Crossref] [PubMed]
- Yokoyama N, Yin D, Malbon CC. Abundance, complexation, and trafficking of Wnt/beta-catenin signaling elements in response to Wnt3a. J Mol Signal 2007;2:11. [Crossref] [PubMed]
- Wlodarchak N, Xing Y. PP2A as a master regulator of the cell cycle. Crit Rev Biochem Mol Biol 2016;51:162-84. [Crossref] [PubMed]
- Erickson RP. Genes, Environment, and Orofacial Clefting: N-Acetyltransferase and Folic Acid. J Craniofac Surg 2010;21:1384-7. [Crossref] [PubMed]
- Gitton Y. Evolving maps in craniofacial development. Semin Cell Dev Biol 2010;21:301-8. [Crossref] [PubMed]
- Chauhan BK, Hoover JM, Scanga H, et al. Isolated Sagittal Synostosis in a Boy with Craniofrontonasal Dysplasia and a Novel EFNB1 Mutation. Plast Reconstr Surg Glob Open 2015;3:e427. [Crossref] [PubMed]
- Tal-Saban M, Ornoy A, Parush S. Young Adults With Developmental Coordination Disorder: A Longitudinal Study. Am J Occup Ther 2014;68:307-16. [Crossref] [PubMed]
- Gong SG. Cranial Neural Crest: Migratory Cell Behavior and Regulatory Networks. Exp Cell Res 2014;325:90-5. [Crossref] [PubMed]
- Simoes-Costa M, Bronner ME. Reprogramming of avian neural crest axial identity and cell fate. Science 2016;352:1570-3. [Crossref] [PubMed]
- Dash S, Trainor P. The development, patterning and evolution of neural crest cell differentiation into cartilage and bone. Bone 2020;137:115409. [Crossref] [PubMed]
- Schoen C, Glennon JC, Abghari S, et al. Differential microRNA expression in cultured palatal fibroblasts from infants with cleft palate and controls. Eur J Orthod 2018;40:90-6. [Crossref] [PubMed]
- Cobourne MT. The complex genetics of cleft lip and palate. Eur J Orthod 2004;26:7-16. [Crossref] [PubMed]
- Kohli SS, Kohli VS. A comprehensive review of the genetic basis of cleft lip and palate. J Oral Maxillofac Pathol 2012;16:64-72. [Crossref] [PubMed]
- Sangodkar J, Farrington CC, McClinch K, et al. All roads lead to PP2A: exploiting the therapeutic potential of this phosphatase. FEBS J 2016;283:1004-24. [Crossref] [PubMed]
- Matthes F, Hettich MM, Schilling J, et al. Inhibition of the MID1 protein complex: a novel approach targeting APP protein synthesis. Cell Death Discovery 2018;4:4. [Crossref] [PubMed]
- Monteiro O, Chen C, Bingham R, et al. Pharmacological disruption of the MID1/a4 interaction reduces mutant Huntingtin levels in primary neuronal cultures. Neurosci Lett 2018;673:44-50. [Crossref] [PubMed]
- Abramyan J, Richman JM. Craniofacial development: discoveries made in the chicken embryo. Int J Dev Biol 2018;62:97-107. [Crossref] [PubMed]
- Howard S, Deroo T, Fujita Y, et al. A positive role of cadherin in Wnt/β-catenin signalling during epithelial-mesenchymal transition. PLoS One 2011;6:e23899. [Crossref] [PubMed]
- Basu S, Cheriyamundath S, Ben-Ze'ev A. Cell–cell adhesion: linking Wnt/β-catenin signaling with partial EMT and stemness traits in tumorigenesis. F1000 Research 2018;7:1488. [Crossref] [PubMed]
- Martyn I, Brivanlou AH, Siggia ED. A wave of WNT signaling balanced by secreted inhibitors controls primitive streak formation in micropattern colonies of human embryonic stem cells. Development 2019;146:dev.172791.
- Kosnopfel C, Sinnberg T, Sauer B, et al. YB-1 Expression and Phosphorylation Regulate Tumorigenicity and Invasiveness in Melanoma by Influencing EMT. Molecular Cancer Research Mcr 2018;16:1149-60. [Crossref] [PubMed]
- Geetha-Loganathan P, Nimmagadda S, Fu K, et al. Avian facial morphogenesis is regulated by c-Jun N-terminal kinase/planar cell polarity (JNK/PCP) wingless-related (WNT) signaling. J Biol Chem 2014;289:24153-67. [Crossref] [PubMed]


