
Transfer RNA-derived small RNAs: potential applications as novel biomarkers for disease diagnosis and prognosis
Introduction
Transfer RNA-derived small RNAs (tsRNAs) are a type of small RNA fragments produced by a given nuclease (such as Dicer and ANG) cutting into the tRNA ring in a given cell/tissue or under specific conditions, such as cell stress (1). tsRNAs originate from precursor or mature transfer RNAs which are cut precisely by enzymes (2), and can be mainly divided into two types based on the cleavage sites (3): tRNA-derived fragments (tRFs) and tRNA-derived stress-induced RNAs (tiRNAs, also known as tRNA halves) (1). tRFs are derived from mature or primary tRNAs that are 14–30 nt in length (dividing into 1-tRFs, 2-tRFs, 3-tRFs, 5-tRFs, and i-tRFs according to the sources), while tiRNAs induced by stress or starvation are produced by specific cleavage at the mature tRNA anticodon loop of over 31 nt (including 5'- and 3'- fragments, named 5'-tiRNAs and 3'-tiRNAs, respectively) (1,3,4). Moreover, tiRNAs are mainly located in the cytoplasm, while the location of tRFs remains controversial due to the variations in previous experimental approaches. However, both tiRNAs and tRFs are delivered into the peripheral blood through the circulatory system and can also be detected by RNA high-throughput sequencing analysis and validated by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR).
A novel optimized bioinformatic tool, SPORTS1.0 can annotate and profile canonical tsRNAs from small RNA (sRNA) sequencing data across a wide range of species (5). According to nucleotide mismatches within sRNAs, SPORTS1.0 can predict potential RNA modification sites and help further understand tsRNA functions. Additionally, databases, such as MINTbase and tRFdb, have been used in the biological analysis of the tsRNAs (6-8).
tsRNAs have been found to exhibit important physiological features in various vital activities, and possess different biological functions depending on their sub-type. tsRNAs are commonly known to regulate gene expression and epigenetic inheritance (1,3). Recent studies established tsRNAs as widely expressed in a variety of mammalian tissues and organs (9). It has also been revealed that tsRNAs, similar to miRNAs, may regulate mRNA stability through binding mechanisms (9-11). tsRNAs have exhibited higher stability (12) and increased expression compared to miRNAs in mouse serum (13). Sharma et al. showed the specificity in testis and epididymis during sperm maturation and fertilization (14), while Karaiskos et al. observed an age-dependent change in tRF expression of rat brain tissues (15). In particular, 3' tRFs expression increased with age, while 5' tRFs showed inconsistent distribution (15). Therefore, tsRNAs are potential candidates for biomarkers across a wide range of clinical indications.
Recently, tsRNAs have been implicated in various diseases, such as cancer, neurological disorders, viral infection, and metabolic diseases. Although their functions are still poorly understood, the abnormal expressions of tRFs and tiRNAs have been extensively observed, with their potential as biomarkers being a target of near-constant research. To identify dysregulated tsRNAs, recent studies have performed high-throughput sequencing analysis and qRT-PCR validation. In this review, we summarize the dysregulated expressions of tsRNAs, and discuss the possibility of tsRNAs as biomarkers in cancer (Figure 1) and other diseases (Figure 2).
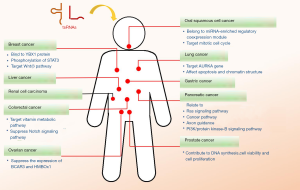
tsRNAs as biomarkers in cancer
A large number of studies have demonstrated that tsRNAs may be ideal biomarkers for cancers of the breast, ovaries, lung, prostate, colorectum, and other organs (Figure 1).
Breast cancer
MiRNAs are consistently reported to be associated with cancer growth, metastasis, and invasion. For instance, recent studies have revealed that “miRNA-like” tRFs in breast cancer are strongly associated with tumor progression (16,17), while markedly altered tRFs (5' tRNA halves) found in the serum of breast cancer patients were significantly associated with clinicopathologic characteristics and subtypes of breast cancer (18).
tRFs have also exhibited tumor-suppressive behavior in breast tissue. Goodarzi et al. demonstrated that a class of tRFs inhibited the development of breast cancer and metastasis (19) by binding to oncogenic protein YBX1 instead of pro-oncogenic transcripts. Also, the circulating expression of tDR-7816, which could influence the xenobiotic metabolic processes to promote oncogenesis of cancer, was validated as a promising biomarker for the diagnosis of patients with early non triple-negative breast cancer (20). Using high-throughput sequencing and bioinformatics analyses, Cui et al. discovered that tDR-0009 and tDR-7336 expression, derived from tRNA (Gly), were increased in hypoxia-treated triple-negative breast cancer cell lines, and were predicted to be involved in the regulation of hypoxia-induced chemoresistance through STAT3 activation (21). Furthermore, Wang et al. proposed three tRFs (upregulated: tRF-32-XSXMSL73VL4YK; downregulated: tRF-32-Q99P9P9NH57SJ and tRF-17-79MP9PP) as potential novel diagnostic biomarkers of breast cancer (22), after revealing their significant expression levels in breast cancer tissues.
The upregulation of tsRNA-26576 in the tissues of breast cancer patients has been associated with the inhibition of cancer cell apoptosis, and the promotion of cancer cell growth, migration, and invasion (23). Moreover, the expression of two tumor suppressor genes, SPEN and FAT4, was found to be increased following treatment with tsRNA-26576 knockdown (23). In addition, sex hormone-dependent tRNA-derived RNA (SHOT-RNA)s were abundantly expressed in human breast cancer cells, and 5'-SHOT-RNAs could enhance tumor cell proliferation (24). Moreover, two novel internal tRFs (i-tRF), tRF-30-JZOYJE22RR33 and tRF-27-ZDXPHO53KSN were reported to be upregulated in metastatic HER-2 positive breast cancer patients and associated with trastuzumab drug resistance and poor prognosis (25). Therefore, the association of tsRNAs and tumor progression supports their potential as clinical biomarkers for breast cancer treatment. The strong correlation between tsRNR expression and cancer development makes them ideal targets for future treatment applications, and they may have further potential in evaluating the therapeutic effects of these treatments.
The abnormal activation of the Wnt/β-Catenin pathway has been significantly associated with tumorigenesis (26). A recent study demonstrated the inhibition of the Wnt/β-Catenin pathway by 5'-tiRNA(Val) overexpression in both serum and tumor tissues of breast cancer patients. In this study, 5'-tiRNA(Val) was significantly downregulated in breast cancer tissue (27). The inhibitory mechanism adopted by 5'-tiRNA(Val) was facilitated by targeting FZD3. tRF3E, another newly discovered tRF derived from tRNA(Glu), was downregulated in the plasma of patients with HER2-positive breast cancer compared to healthy controls (28). In this study, authors showed that tRF3E plays an essential role as a tumor-suppressor through targeting the RNA-binding protein, nucleolin.
Ovarian cancer
Ovarian cancer is another sex hormone-associated cancer (29). A study revealed that tRF-03357 and tRF-03358 were significantly increased in the serum of high-grade ovarian cancer patients and cells. In particular, tRF-03357 promoted cancer cell proliferation, migration, and invasion via inhibiting the tumor-suppressive factor, HMBOX1 (30). In the contrast, Zhou et al. reported that tRF5-Glu could directly target breast cancer anti-estrogen resistance 3 (BCAR3) mRNA and disturb its expression in ovarian cancer, resulting in the inhibition of ovarian cancer cell proliferation (31). Therefore, tRFs should be strongly considered as potential diagnostic biomarkers and therapeutic targets for future clinical applications in ovarian cancer.
Lung cancer
Lung cancer is responsible for the highest cancer mortality rate over the past few decades and is one of the most common types of malignant tumors worldwide (32). Pekarsky et al. identified two novel tiRNAs, ts-4521 and ts-3676, as potentially exhibiting anti-tumor functions in lung cancer and chronic lymphocytic leukemia (CLL). These tiRNAs were downregulated in lung cancer tissue and CLL (33). Both tiRNAs were reported to be involved in apoptosis and chromatin structure, indicating their potential role in regulating tumor cell growth and survival of lung cancer cells (34). Moreover, it also has been reported that tRF-Leu-CAG is upregulated in human non‐small cell lung cancer, particularly in the late stages of the disease, and is associated with cell cycle progression by targeting AURKA (35). These results indicate that tsRNAs play an important role in tumor progression and further support dysregulated tsRNAs as promising biomarkers for lung cancer diagnosis.
Prostate cancer
Prostate cancer is the second most common cancer in men (32). As previously mentioned, Honda et al. observed enriched SHOT-RNA expression in prostate cancer and enhanced tumor proliferation (24). In a previous study, tRF-1001 was required for cell proliferation and highly expressed in prostate cancer cell lines compared to tissues (36). tRF-1001 knockdown treatment resulted in the inhibition of DNA synthesis, cell viability, and cell proliferation by arresting cancer cells at the G2 phase of the cell cycle (36). Hormone-dependent tiRNAs were reported to be upregulated in prostate cancer patients (37). The high level of these tiRNAs was strongly associated with adverse clinicopathological parameters, demonstrating that those tiRNAs could function as prognostic predictors for prostate cancer patients (37). Moreover, Olvedy et al. analyzed the expression profiles of tRFs in prostate cancer samples and found tRF-544 from tRNA(Phe)-GAA to be downregulated and tRF-315 from tRNA(Lys)-CTT to be upregulated in prostate cancer tissues compared to healthy tissues (38). The ratio of tRF-315/tRF-544 was positively associated with poorer progression-free survival and shorter period to disease relapse, indicating that the tRF-315 to tRF-544 ratio might be a potential clinical biomarker for the tumor progression of prostate cancer (38).
Colorectal cancer
Colorectal cancer is one of the leading common cancers which causes 600,000 deaths globally every year (39). Through comprehensive analysis, 16 tRFs and 5 key miRNAs were identified as being significantly altered in colon cancer compared to matched peritumor tissues (40). Target mRNAs of the 16 tRFs and 5 miRNAs were primarily involved in vitamin metabolic pathways and the cyclic guanine monophosphate-protein kinase G signaling pathway, indicating their potential roles in colon cancer development (40). It has been demonstrated that tRF/miR-1280 is derived from both tRNA (Leu) and pre-miRNA-1280, and serves as a critical regulator of cancer growth in colorectal cancer cells (41,42). Li et al. showed that 5'-tiRNA-Val was upregulated in colorectal cancer and could be involved in angiogenin (ANG)-mediated tumor metastasis (43). Further mechanism-focused investigations have revealed that tRF/miR-1280 to be less expressed in colorectal cancer and targeted Notch ligand JAG2 (42). These studies suggest that tsRNAs might represent a potential target biomarker in the clinical prediction of colorectal cancer diagnosis and prognosis.
Other cancers
In addition to the cancers detailed above, recent studies have found that tiRNAs or tRFs could potentially serve as biomarkers for the clinical diagnosis of other cancers. Jin et al. investigated the expression profile of tRFs in clinical samples of pancreatic cancer. The target genes of four validated tRFs exhibiting differential expressions (AS-tDR-000064, AS-tDR-000069, AS-tDR-000102, and AS-tDR-001291) were mostly enriched in the Ras signaling, cancer, axon guidance, and PI3K/protein kinase-B signaling pathway according to Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (44). This demonstrates that tsRNAs could also contribute to pancreatic tumorigenesis and might be used as a clinical tool for the diagnosis and prognosis of pancreatic cancer (44). Zhu et al. also demonstrated that plasma exosome tsRNAs could be a potential novel “liquid biopsy” biomarkers for liver cancer diagnosis (45). Four tsRNAs were reported to be increased in plasma exosomes of liver cancer patients compared to healthy controls, supporting the diagnostic value of plasma exosome tsRNAs in cancer (45). Furthermore, tiRNA-5034-GluTTC-2 was found to be downregulated in tissues and plasma of gastric cancer, and patients with higher tiRNA-5034-GluTTC-2 expression exhibited a longer overall curve compared to those with low expression (46).
Previous studies have also demonstrated that tiRNAs were dysregulated in clear cell renal cell carcinoma (RCC). In particular, 5'-tRNA halves were downregulated in tissues and serum RCC patients, and thus may act as potential biomarkers for diagnosis and prognosis (47,48). However, the mechanisms of these tsRNAs remain to be further investigated.
Among patients with head and neck squamous cell carcinoma, the circulation levels of 5'-tiRNAs were significantly dysregulated in serum of cancer patients (49). Interestingly, tsRNAs might have different expression patterns in circulatory and tumor tissues. For instance, 5' tRNA-Val-CAC-2-1 was downregulated in the serum of oral squamous cell carcinoma patients but was upregulated in tumors compared to healthy adjacent tissue (50). Another, study revealed that 5' tRNA-Val-CAC-2-1 half belonged to a miRNA-enriched regulatory co-expression module and targeted genes involved in the regulation of mitosis (50).
In summary, some tsRNAs are highly expressed in tumor tissues and serum/plasma of cancer patients. They moderate cell growth by directly or indirectly targeting oncogenic proteins or pathways, and have been strongly associated with tumor growth, invasion, and metastasis. However, some tsRNAs showed lower expression in cancer patients and were reported to elicit tumor-suppressive effects. Thus, dysregulated tsRNAs could be used as novel biomarkers for the diagnosis and prognosis of different cancers.
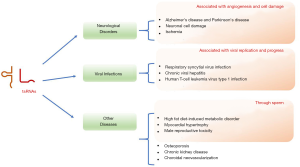
Neurological disorders
Neurological diseases are structural and functional disorders of the nervous system (51). In the senescence-accelerated mouse prone 8 (SAMP8) model (a model of Alzheimer’s disease and Parkinson’s disease), the expression of tsRNAs were significantly altered in the brain of SAMP8 mice, with dysregulated tsRNAs strongly associated with impaired synapse formation and disrupted synaptic vesicle cycle pathways (52). Li et al. reported two tsRNAs from tRNA(Val)-CAC and tRNA(Gly)-GCC, which were abundantly expressed in the rat brain ischemia model, the mouse hindlimb ischemia model, and the cellular hypoxia model (53). Functional analyses revealed that these upregulated tsRNAs could suppress angiogenesis and could be potential therapeutic targets (53). Under cell stress, tRNAs were found to be cleaved into small tiRNAs in rat neurons (54), and the levels of generated tiRNAs have been strongly associated with the severity of cell damage, indicating tiRNAs could function as a potential biomarker of neuron damage (54,55). Furthermore, tiRNA levels were observed to be increased in neurons soon after reperfusion, while its levels were significantly reduced in cells treated with minocycline, a neuroprotective antibiotic preventing reperfusion injury. This highlights the potential use of tiRNAs as indicators of treatment effect in ischemic injury (55).
ANG mutants have been associated with increased risk for Parkinson’s disease and amyotrophic lateral sclerosis (56,57). ANG-induced tsRNAs could therefore indicate the development and diagnosis of Parkinson’s disease. Magee et al. showed that the expressions of tsRNAs in the prefrontal cortex, cerebrospinal fluid, and serum samples, could distinguish Parkinson’s disease patients from healthy controls, indicating a potential role for tsRNAs as reliable biomarkers and predictive tools of Parkinson’s disease (58).
These studies demonstrated that aberrant tsRNA metabolism indicating dysregulation of tsRNAs was potentially associated with neurodegeneration. Further studies are needed to reveal the diagnostic and prognostic capability of tsRNAs.
Viral infections
To investigate the global expression profile of tsRNAs in respiratory syncytial virus (RSV)-infected airway epithelial cells, Wang et al. found a robust induction of tRFs derived from tRNA cleavages (59). Among them, tRF5-GluCTC was generated by ANG and promoted RSV replication (59). A further study revealed that RSV replication was regulated by suppressing the apolipoprotein E receptor 2, a known anti-RSV protein and target of tRF-GluCTC (60). Moreover, two novel RSV-induced tsRNAs (tRF5-GlyCCC and tRF5-CysGCA) were markedly increased with RSV infection, and promoted RSV replication (61). Inhibition of these two tsRNAs with antisense treatment suppressed RSV proliferation and virus-associated inflammation, implicating anti-tsRNAs as a promising therapy for the control of RSV (61). Another study found 30–35 nt 5'-tiRNAs to be abundantly expressed in healthy liver tissues compared to cancerous liver tissues, and to be increased during infection with chronic viral hepatitis (62). Additional deep sequencing analysis showed that tRFs were more abundant in human T-cell leukemia virus type 1 (HTLV-1) infected T cells than in healthy counterparts (63). Among these tRFs, tRF-1001, which was upregulated in prostate cancer cells (36), was found to be increased in HTLV-1–infected cells (63). Another tRF, tRF-3019, was shown to be upregulated in infected cells and viral particles and was observed functioning as a primer for HTLV-1 reverse transcriptase, indicating that tRF-3019 might be critically involved in HTLV-1 replication (63). The studies, taken together, suggest that the tsRNA expression can serve as a biomarker for the diagnosis and prognosis of viral infection. Moreover, the strong association between tsRNA expression and viral replication suggests that tsRNAs may play an important role in the treatment and management of viral infections.
Other diseases
Numerous studies reported the roles of tsRNAs in other diseases. Accumulating evidence concerning the paternal hereditary pattern of diet-induced metabolic disorder has suggested the involvement of sperm in transgenerational epigenetic inheritance (64-66). A novel class of tsRNA derived from 5'tRNAs was first discovered in mammal mature sperm (67). Although detailed molecular mechanisms and functions of sperm tsRNAs remain unknown, recent studies have shown sperm tsRNAs to be significantly elevated in mice with high-fat diet (HFD)-induced metabolic disorders (68), or a low-protein diet (14). Further investigation has found tRNA-Gly-GCC expression to be decreased in the testis but increased in the sperm. tRNA-Gly-GCC also modulated the activity of endogenous retroelements in the preimplantation embryo (14). A parental HFD may also influence the development of metabolic disorders in their offspring through transmission of specific tsRNAs from the sperm. This results in impaired glucose tolerance and body weight loss across two generations (69). Another study found that the sperm tRFs from obese men to be significantly altered (70). Moreover, a novel multiple generation mouse model demonstrated that maternal HFD could induce obesogenic phenotypes across three generations through sperm tsRNAs, indicating its lasting effect on transgenerational epigenetic inheritance (71). Sperm RNA modifications may also be intergenerationally transmitted. Zhang et al. demonstrated that a methyltransferase, DNMT2, mediated paternal HFD-induced metabolic disorder transmission by altering the sperm tsRNA expression profile (72). Environmental exposure can also promote tsRNAs changes in the sperm, inducing a higher incidence of various diseases in offspring (73). In a myocardial hypertrophy rat model, small tRFs were found to be highly expressed in hypertrophic parental rat sperm, resulting in increased hypertrophic markers, fibrosis, and apoptosis in the hearts of offspring (74). In another rat model, sperm tsRNAs were found to be predictive and sensitive biomarkers of male reproductive toxicity exposed to ethylene glycol monomethyl ether (75). These studies indicate the possible function of sperm tsRNAs as potential vectors that transmit parental diet-induced metabolic disorders, and further investigations are required to demonstrate the feasibility of sperm tsRNAs as potential biomarkers for the diagnosis of hereditary diseases in subsequent generations.
In addition to tsRNAs, three plasma exosomal tRFs (tRF-18, tRF-25, tRF-38), derived from tRNA (Val) were shown to be significantly increased in patients with osteoporosis compared to healthy controls (76). Two urinary exosomal tRFs, tRF-Val-TAG and tRF-Leu-AAC, which originate from kidney-related cells, were found to be significantly decreased in the patients with chronic kidney disease (77). The association between these tRFs and reduction of the glomerular filtration rate needs to be further investigated. In a recent study, we reported 72 tsRNAs that were significantly altered in a laser-induced choroidal neovascularization mouse model. Further studies by Gene Ontology (GO) and KEGG pathway analyses revealed that the target mRNAs of the altered tsRNAs were most enriched in the regulation of immune system processes, extracellular region, core promoter-binding and NOD-like receptor signaling pathway (78).
Conclusions
tsRNAs are novel, non-coding RNAs which are garnering increasing focus within the research community, but their functions and mechanisms remain largely unknown. This review provides a consolidated and compelling view of tsRNAs, including tRFs and tiRNAs, as novel biomarkers for various diseases including cancer, neurological disorders, viral infections, and metabolic diseases and angiogenesis-related diseases. The regulation of tsRNAs is, to some extent, similar to miRNAs in regards to the related physiological and pathological processes. However, the stability and higher expression levels exhibited by tsRNAs place them as ideal diagnostic and prognostic biomarkers. In order to fully utilize tsRNAs in clinical settings across various diseases, their functions and mechanisms need to be imminently clarified via further investigation.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (No. 81800855), Natural Science Foundation of Hunan Province (No. 2018JJ3765), Changsha Science and Technology Project (No. kq1907075) and Department of Science and Technology, Hunan (No. 2015TP2007).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2797). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shen Y, Yu X, Zhu L, et al. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J Mol Med (Berl) 2018;96:1167-76. [Crossref] [PubMed]
- Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet 2014;5:171. [Crossref] [PubMed]
- Li S, Xu Z, Sheng J. tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes (Basel) 2018;9:246. [Crossref] [PubMed]
- Zhu L, Ge J, Li T, et al. tRNA-derived fragments and tRNA halves: The new players in cancers. Cancer Lett 2019;452:31-7. [Crossref] [PubMed]
- Shi J, Ko E-A, Sanders KM, et al. SPORTS1.0: A Tool for Annotating and Profiling Non-coding RNAs Optimized for rRNA- and tRNA-derived Small RNAs. Genomics Proteomics Bioinformatics 2018;16:144-51. [Crossref] [PubMed]
- Pliatsika V, Loher P, Telonis AG, et al. MINTbase: a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics 2016;32:2481-9. [Crossref] [PubMed]
- Pliatsika V, Loher P, Magee R, et al. MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res 2018;46:D152-9. [Crossref] [PubMed]
- Kumar P, Mudunuri SB, Anaya J, et al. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res 2015;43:D141-5. [Crossref] [PubMed]
- Kumar P, Anaya J, Mudunuri SB, et al. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 2014;12:78. [Crossref] [PubMed]
- Shigematsu M, Kirino Y. tRNA-Derived Short Non-coding RNA as Interacting Partners of Argonaute Proteins. Gene Regul Syst Bio 2015;9:27-33. [Crossref] [PubMed]
- Kim HK, Fuchs G, Wang S, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 2017;552:57-62. [Crossref] [PubMed]
- Zhang Y, Zhang Y, Shi J, et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J Mol Cell Biol 2014;6:172-4. [Crossref] [PubMed]
- Victoria B, Dhahbi JM, Nunez Lopez YO, et al. Circulating microRNA signature of genotype-by-age interactions in the long-lived Ames dwarf mouse. Aging Cell 2015;14:1055-66. [Crossref] [PubMed]
- Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016;351:391-6. [Crossref] [PubMed]
- Karaiskos S, Grigoriev A. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Res 2016;5:ISCB Comm J-2758.
- Guzman N, Agarwal K, Asthagiri D, et al. Breast Cancer-Specific miR Signature Unique to Extracellular Vesicles Includes "microRNA-like" tRNA Fragments. Mol Cancer Res 2015;13:891-901. [Crossref] [PubMed]
- Telonis AG, Rigoutsos I. Race Disparities in the Contribution of miRNA Isoforms and tRNA-Derived Fragments to Triple-Negative Breast Cancer. Cancer Res 2018;78:1140-54. [Crossref] [PubMed]
- Dhahbi JM, Spindler SR, Atamna H, et al. Deep Sequencing of Serum Small RNAs Identifies Patterns of 5' tRNA Half and YRNA Fragment Expression Associated with Breast Cancer. Biomark Cancer 2014;6:37-47. [Crossref] [PubMed]
- Goodarzi H, Liu XH, Nguyen HCB, et al. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015;161:790-802. [Crossref] [PubMed]
- Huang Y, Ge H, Zheng MJ, et al. Serum tRNA-derived fragments (tRFs) as potential candidates for diagnosis of nontriple negative breast cancer. J Cell Physiol:16.
- Cui Y, Huang Y, Wu X, et al. Hypoxia-induced tRNA-derived fragments, novel regulatory factor for doxorubicin resistance in triple-negative breast cancer. J Cell Physiol 2019;234:8740-51. [Crossref] [PubMed]
- Wang X, Yang Y, Tan X, et al. Identification of tRNA-Derived Fragments Expression Profile in Breast Cancer Tissues. Curr Genomics 2019;20:199-213. [Crossref] [PubMed]
- Zhou J, Wan F, Wang YK, et al. Small RNA sequencing reveals a novel tsRNA-26576 mediating tumorigenesis of breast cancer. Cancer Manag Res. 2019;11:3945-56. [Crossref] [PubMed]
- Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci USA 2015;112:E3816-25. [Crossref] [PubMed]
- Sun C, Yang F, Zhang Y, et al. tRNA-Derived Fragments as Novel Predictive Biomarkers for Trastuzumab-Resistant Breast Cancer. Cell Physiol Biochem 2018;49:419-31. [Crossref] [PubMed]
- Clevers H, Nusse R. Wnt/beta-Catenin Signaling and Disease. Cell 2012;149:1192-205. [Crossref] [PubMed]
- Mo D, Jiang P, Yang Y, et al. A tRNA fragment, 5 ‘-tiRNA(val), suppresses the Wnt/beta-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer Lett 2019;457:60-73. [Crossref] [PubMed]
- Falconi M, Giangrossi M, Zabaleta ME, et al. A novel 3'-tRNAGlu-derived fragment acts as a tumor suppressor in breast cancer by targeting nucleolin. FASEB J 2019;33:13228-40. [Crossref] [PubMed]
- Chuffa LGD, Lupi LA, Costa AB, et al. The role of sex hormones and steroid receptors on female reproductive cancers. Steroids 2017;118:93-108. [Crossref] [PubMed]
- Zhang M, Li F, Wang J, et al. tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer. Oncotargets and Therapy 2019;12:6371-83. [Crossref] [PubMed]
- Zhou K, Diebel KW, Holy J, et al. A tRNA fragment, tRF5-Glu, regulates BCAR3 expression and proliferation in ovarian cancer cells. Oncotarget 2017;8:95377-91. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Pekarsky Y, Balatti V, Palamarchuk A, et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci USA 2016;113:5071-6. [Crossref] [PubMed]
- Balatti V, Nigita G, Veneziano D, et al. tsRNA signatures in cancer. Proc Natl Acad Sci USA 2017;114:8071-6. [Crossref] [PubMed]
- Shao Y, Sun QL, Liu XM, et al. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des 2017;90:730-8. [Crossref] [PubMed]
- Lee YS, Shibata Y, Malhotra A, et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 2009;23:2639-49. [Crossref] [PubMed]
- Zhao C, Tolkach Y, Schmidt D, et al. tRNA-halves are prognostic biomarkers for patients with prostate cancer. Urol Oncol 2018;36:503.e1-503.e7. [Crossref] [PubMed]
- Olvedy M, Scaravilli M, Hoogstrate Y, et al. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget 2016;7:24766-77. [Crossref] [PubMed]
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014;383:1490-502. [Crossref] [PubMed]
- Xiong W, Wang XL, Cai XY, et al. Identification of tRNA-derived fragments in colon cancer by comprehensive small RNA sequencing. Oncol Rep 2019;42:735-44. [PubMed]
- Schopman NCT, Heynen S, Haasnoot J, et al. A miRNA-tRNA mix-up tRNA origin of proposed miRNA. Rna Biology 2010;7:573-6. [Crossref] [PubMed]
- Huang B, Yang H, Cheng X, et al. tRF/miR-1280 Suppresses Stem Cell-like Cells and Metastasis in Colorectal Cancer. Cancer Res 2017;77:3194-206. [Crossref] [PubMed]
- Li S, Shi X, Chen M, et al. Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int J Cancer 2019;145:1395-407. [Crossref] [PubMed]
- Jin L, Zhu C, Qin X. Expression profile of tRNA-derived fragments in pancreatic cancer. Oncol Lett 2019;18:3104-14. [PubMed]
- Zhu L, Li J, Gong Y, et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer 2019;18:74. [Crossref] [PubMed]
- Zhu L, Li T, Shen Y, et al. Using tRNA halves as novel biomarkers for the diagnosis of gastric cancer. Cancer Biomark 2019;25:169-76. [Crossref] [PubMed]
- Zhao C, Tolkach Y, Schmidt D, et al. 5 ‘-tRNA Halves are Dysregulated in Clear Cell Renal Cell Carcinoma. J Urol 2018;199:378-83. [Crossref] [PubMed]
- Nientiedt M, Deng M, Schmidt D, et al. Identification of aberrant tRNA-halves expression patterns in clear cell renal cell carcinoma. Sci Rep 2016;6:37158. [Crossref] [PubMed]
- Martinez B, Dhahbi J, Lopez Y, et al. Circulating small non coding RNA signature in head and neck squamous cell carcinoma. Oncotarget 2015;6:19246-63. [Crossref] [PubMed]
- Dhahbi J, Lopez YON, Schneider A, et al. Profiling of tRNA Halves and YRNA Fragments in Serum and Tissue From Oral Squamous Cell Carcinoma Patients Identify Key Role of 5 ‘ tRNA-Val-CAC-2-1 Half. Front Oncol 2019;9:959. [Crossref] [PubMed]
- Faghihi MA, Mottagui-Tobor S, Wahlestedt C. Genetics of neurological disorders. Expert Rev Mol Diagn 2004;4:317-32. [Crossref] [PubMed]
- Zhang S, Li H, Zheng L, et al. Identification of functional tRNA-derived fragments in senescence-accelerated mouse prone 8 brain. Aging (Albany NY) 2019;11:10485-98. [Crossref] [PubMed]
- Li Q, Hu B, Hu G-W, et al. tRNA-Derived Small Non-Coding RNAs in Response to Ischemia Inhibit Angiogenesis. Sci Rep 2016;6:20850. [Crossref] [PubMed]
- Elkordy A, Mishima E, Niizuma K, et al. Stress-induced tRNA cleavage and tiRNA generation in rat neuronal PC12 cells. J Neurochem 2018;146:560-9. [Crossref] [PubMed]
- Elkordy A, Rashad S, Shehabeldeen H, et al. tiRNAs as a novel biomarker for cell damage assessment in in vitro ischemia-reperfusion model in rat neuronal PC12 cells. Brain Res 2019;1714:8-17. [Crossref] [PubMed]
- Bradshaw WJ, Rehman S, Pham TTK, et al. Structural insights into human angiogenin variants implicated in Parkinson’s disease and Amyotrophic Lateral Sclerosis. Sci Rep 2017;7:41996. [Crossref] [PubMed]
- van Es MA, Schelhaas HJ, van Vught PWJ, et al. Angiogenin variants in Parkinson disease and amyotrophic lateral sclerosis. Ann Neurol 2011;70:964-73. [Crossref] [PubMed]
- Magee R, Londin E, Rigoutsos I. TRNA-derived fragments as sex-dependent circulating candidate biomarkers for Parkinson’s disease. Parkinsonism Relat Disord 2019;65:203-9. [Crossref] [PubMed]
- Wang Q, Lee I, Ren J, et al. Identification and Functional Characterization of tRNA-derived RNA Fragments (tRFs) in Respiratory Syncytial Virus Infection. Mol Ther 2013;21:368-79. [Crossref] [PubMed]
- Deng J, Ptashkin R, Chen Y, et al. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Mol Ther 2015;23:1622-9. [Crossref] [PubMed]
- Zhou J, Liu S, Chen Y, et al. Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J Gen Virol 2017;98:1600-10. [Crossref] [PubMed]
- Selitsky SR, Baran-Gale J, Honda M, et al. Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Sci Rep 2015;5:7675. [Crossref] [PubMed]
- Ruggero K, Guffanti A, Corradin A, et al. Small Noncoding RNAs in Cells Transformed by Human T-Cell Leukemia Virus Type 1: a Role for a tRNA Fragment as a Primer for Reverse Transcriptase. J Virol 2014;88:3612-22. [Crossref] [PubMed]
- Carone BR, Fauquier L, Habib N, et al. Paternally Induced Transgenerational Environmental Reprogramming of Metabolic Gene Expression in Mammals. Cell 2010;143:1084-96. [Crossref] [PubMed]
- Ng SF, Lin RCY, Laybutt DR, et al. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature 2010;467:963-6. [Crossref] [PubMed]
- Rando OJ. Daddy Issues: Paternal Effects on Phenotype. Cell 2012;151:702-8. [Crossref] [PubMed]
- Peng H, Shi J, Zhang Y, et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res 2012;22:1609-12. [Crossref] [PubMed]
- Chen Q, Yan MH, Cao ZH, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016;351:397-400. [Crossref] [PubMed]
- de Castro Barbosa T, Ingerslev LR, Alm PS, et al. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol Metab 2015;5:184-97. [Crossref] [PubMed]
- Donkin I, Versteyhe S, Ingerslev LR, et al. Obesity and Bariatric Surgery Drive Epigenetic Variation of Spermatozoa in Humans. Cell Metab 2016;23:369-78. [Crossref] [PubMed]
- Sarker G, Sun WF, Rosenkranz D, et al. Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs. Proc Natl Acad Sci U S A 2019;116:10547-56. [Crossref] [PubMed]
- Zhang Y, Zhang X, Shi J, et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol 2018;20:535. [Crossref] [PubMed]
- Schuster A, Skinner MK, Yan W. Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ Epigenet 2016;2:10.
- Shen L, Gan M, Tan Z, et al. A Novel Class of tRNA-Derived Small Non-Coding RNAs Respond to Myocardial Hypertrophy and Contribute to Intergenerational Inheritance. Biomolecules 2018;8:54. [Crossref] [PubMed]
- Stermer AR, Reyes G, Hall SJ, et al. Small RNAs in Rat Sperm Are a Predictive and Sensitive Biomarker of Exposure to the Testicular Toxicant Ethylene Glycol Monomethyl Ether. Toxicol Sci 2019;169:399-408. [Crossref] [PubMed]
- Zhang Y, Cai F, Liu J, et al. Transfer RNA-derived fragments as potential exosome tRNA-derived fragment biomarkers for osteoporosis. Int J Rheum Dis 2018;21:1659-69. [Crossref] [PubMed]
- Khurana R, Ranches G, Schafferer S, et al. Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. Rna 2017;23:142-52. [Crossref] [PubMed]
- Zhang L, Liu S, Wang J-H, et al. Differential Expressions of microRNAs and Transfer RNA-derived Small RNAs: Potential Targets of Choroidal Neovascularization. Curr Eye Res 2019;44:1226-35. [Crossref] [PubMed]


