The relationship between KRAS gene mutation and intestinal flora in tumor tissues of colorectal cancer patients
Introduction
Colorectal cancer is among the most common malignant tumors worldwide. The complex pathogenic process of colorectal cancer in tumor tissues makes this disease particularly difficult to detect and cure at an early stage, hence the increasingly lower 5-year survival rates of patients (1,2). Colorectal cancer mainly involves abdominal organs, though it can metastasize into the lungs or other body tissues. Patients with this disease typically experience symptoms such as abdominal pain and bloody stool and are often diagnosed through digital rectal examination. The majority of patients will remain at middle-advanced stages, and so the therapeutic effect tends to be less favorable. The Kirsten ras (KRAS) gene is highly influential in colorectal cancer (3) and plays a key role in the epidermal growth factor receptor (EGFR) and Ras signaling pathways (4). KRAS gene mutation has a significant influence on the progression and treatment of colorectal cancer (5). Compared to those with wild-type KRAS colorectal cancer, colorectal cancer patients with a mutated KRAS gene exhibit poorer treatment outcomes and accelerated disease progression (6). Evaluating the differences between the patients with wild-type and mutated KRAS colorectal cancer will help to uncover the specific progression mechanism of the disease and guide treatment strategies.
Intestinal microbes are critical for the normal development of the body and the maintenance of intestinal homeostasis. The composition and type of intestinal microbes vary among different diseases, thereby affecting the progression (and even malignancies) of those conditions. This has become an important area of research in recent years (7-9). Being an abdominal tumor, colorectal cancer mainly implicates the digestive tract. The composition and abundance of intestinal flora in patients with colorectal cancer differ substantially from those in healthy individuals, implying an important role of intestinal flora in tumor progression (10).
This study aims to identify the relationship between KRAS gene mutation and intestinal flora and to further elucidate the pathogenesis of colorectal cancer by analyzing the differences in the composition of intestinal microbes between wild-type and mutated KRAS colorectal cancer patients.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5622).
Methods
General information
In the present study, cancer tissue specimens surgically removed from 177 colorectal cancer patients in our hospital were collected. Patients were diagnosed with colorectal cancer by hematoxylin and eosin (HE) staining and histopathological examination of cancer tissue at the Department of Pathology in our hospital. At the same time, the patients’ basic information and general conditions including name, age, sex, tumor stage, and metastasis were recorded. Also, the middle-posterior segments of fresh stools were collected from 30 of these patients and cryopreserved in liquid nitrogen for detecting intestinal flora. All patients were informed of this study and consented to participate in it, with the approval of the Ethics Committee in our hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Study methods
Extraction of deoxyribonucleic acid (DNA) from tissues
The fresh colorectal cancer tissues surgically resected from the patients were taken, sheared into pieces using dissecting scissors, and then ground and stored in centrifugal tubes. Subsequently, 500 µL of lysis buffer, 50 µL of SDS, and 3 µL of protease K was added to the centrifugal tubes and shaken at 60 °C overnight. The next day, the centrifugal tubes were taken out and centrifuged at 7,500 rpm at room temperature for 15 min. After this, the supernatant was collected, and an equivalent volume of isopropanol was added and mixed evenly, and then transferred into spin columns. After centrifugation at 12,000 rpm for 15 min, washing buffer was added to the products and centrifuged at 12,000 rpm for 2 min. Subsequently, the spin columns were placed in new centrifuge tubes, then let to stand at room temperature for 8 min. Thereafter, Tris-EDTA (TE) buffer was added and centrifuged at 12,000 rpm for 1 min. The resulting solution was the DNA of the cancer tissues.
Detection of KRAS gene mutation via real-time fluorescence quantitative polymerase chain reaction (qPCR)
Mutation of the KRAS gene was detected using the real-time fluorescence qPCR kit, and all of the operations were performed strictly according to the manufacturer’s instructions. The total reaction system (50 µL) comprised of 5 µL of DNA templates, 2 µL of KRAS forward primers, 2 µL of KRAS reverse primers, and 41 µL of the mixed reaction system. Negative, positive, and blank controls were set in each group of experiments. Also, the PCR parameters were set as follows: 1× (95 °C for 1 min), 20× (95 °C for 20 s, 60 °C for 25 s, and 72 °C for 25 s) and 35× (92 °C for 30 s, 62 °C for 25 s, and 72 °C for 25 s).
Detection of intestinal flora
Middle-posterior segments of the stools of 30 colorectal cancer patients were sent to TinyGene Bio-Tech (Shanghai) Co., Ltd. for the detection of intestinal microbes. Following this, genomic DNAs in the microbes were extracted, amplified, and a DNA library was constructed and labeled. High-throughput sequencing was then performed using IlluminaMiSeq and Ion PGM to detect the type and relative abundance of microbes in the samples and the related bioinformatics analysis.
Statistical analysis
SPSS 22.0 was used for data statistics and analysis, and enumeration and measurement data were examined using the chi-square test and the t test, respectively. The survival analysis was performed using the Kaplan-Meier method, and the correlation was analyzed using the Pearson method. P<0.05 signified that the difference was statistically significant.
Results
General conditions and KRAS gene mutation in 177 patients
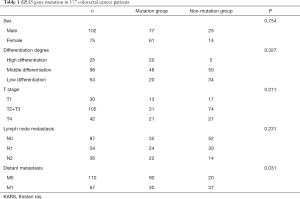
The general conditions and KRAS gene mutation are summarized in Table 1. Both groups of colorectal cancer patients (those with KRAS gene mutation and those with no KRAS gene mutation) exhibited no obvious differences in sex (P=0.734), differentiation degree of tumors (P=0.327), infiltration depth of tumors (P=0.211), and lymph node metastasis (P=0.231). However, there was a statistical difference in the distant metastasis of tumors between the two groups of patients (P=0.031).
Full table
Survival analysis
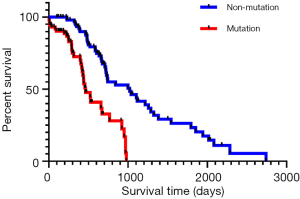
According to the survival analysis results (Figure 1), the mutation group had a poorer prognosis than the non-mutation group (P<0.001). In other words, colorectal cancer patients without KRAS gene mutation showed a better survival prognosis.
Analysis of the differences in the flora between the mutation and non-mutation groups
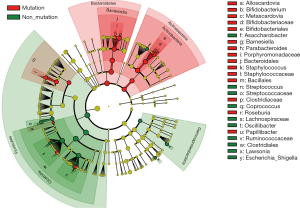
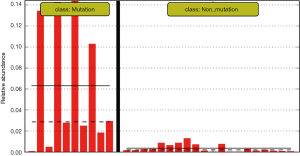
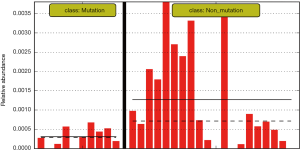
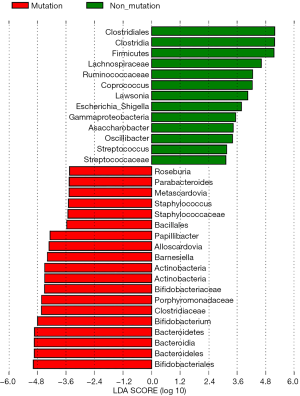
The differences in the intestinal flora between the mutation and non-mutation groups were analyzed using linear discriminant analysis (LDA) Effect size (LEfSe) analysis. It was found that the LDA scores of microbes, such as Roseburia, Parabacteroides, Metascardovia, Staphylococcus, Staphylococcaceae, and Bacillales were significantly higher in the mutation group compared to the non-mutation group, while the LDA scores of Clostridiales, Bacteroidetes, Lachnospiraceae, Coprococcus, and Ruminococcaceae were notably lower in the mutation group compared to the non-mutation group (Figure 2). The cladogram of intestinal flora evolution (Figure 3) shows that the abundance of Bacteroidetes and Actinobacteria in the mutation group was markedly higher than the non-mutation group (Figure 4). Additionally, the non-mutation group had a substantially higher level of intestinal Coriobacteriaceae than the mutation group (Figure 5).
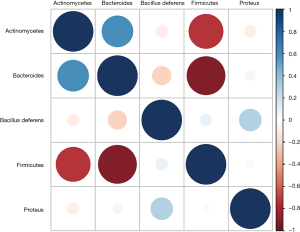
Correlation analysis of intestinal flora
The correlation analysis results of intestinal flora in colorectal cancer patients (Figure 6) revealed that Firmicutes were highly negatively correlated with the abundance of Actinobacteria and Bacteroidetes, while Bacteroidetes were markedly positively associated with the level of Actinobacteria.
Discussion
Colorectal cancer is one of the leading malignant tumors endangering the health of people worldwide. In China, the morbidity and mortality of this disease is increasing annually, and the rate of increase is accelerating. This may be associated with the dietary habits and irregular living arrangements of people.
One of the key features of colorectal cancer is that it is closely related to genetic factors such as gene mutation, gene polymorphism, and chromosome structure or functional variation. The KRAS gene, a crucial molecule in the major intracellular signaling pathways (such as the EGFR and the mitogen activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathways), exerts substantial regulatory effects on multiple tumors. For example, its proteins can promote the growth (11) and metastasis (12,13) of most tumors and serve as the evaluation indexes for chemotherapy in pancreatic cancer (14). The KRAS gene has an elevated mutation rate in some tumors, which may regulate important signaling pathways and protein networks (15). Therefore, the incidence rate of KRAS gene mutation in colorectal cancer patients was studied by our research team, and a correlation between KRAS gene mutation and the distant metastasis of colorectal cancer was found (P=0.031). This suggests that KRAS gene mutation may be associated with the related mechanisms of the distant metastasis of tumors, such as epithelial-mesenchymal transition (EMT), though this remains an area of further study. According to the findings in this study, KRAS gene mutation also dramatically influenced the survival prognosis of colorectal cancer patients (P<0.01), which further supports the significance and the potential key regulatory role of the KRAS gene in this disease, and is consistent with reports in China and beyond.
In CRC, more than 35% patients showed KRAS mutation, and KRAS mutations often appear as single nucleotide variants (16). The most frequent mutation locations were within codon 12 and codon 13 in exon 2, and the most common patterns are G12D, G12A, G12R, G12C, G12S, G12V and G13D (17,18).When mutated, the intrinsic GTPase activity of KRAS would be disturbed, thus leading to the continuous of GTP-bound state and the accumulation of KRAS protein, which would then start the downstream pro-proliferative pathways (19).
The composition and abundance of intestinal microbes, which are important components in the digestive tracts of healthy individuals, have been reported to be associated with several diseases. They have thus become areas of major research in China and internationally in recent years (20,21). According to reports, intestinal flora affects the growth and metastasis of multiple tumors (22), including gastric cancer (23), colorectal cancer (24), breast cancer (25) and liver cancer (26). The present study linked KRAS gene mutation with intestinal flora in colorectal cancer for the first time, and identified differences in intestinal flora between mutation and non-mutation groups, indicating that some intestinal microbes with markedly different abundance may modulate the proliferation, invasion, and metastasis of tumors. Moreover, some microbes were reported in this study for the first time in China (and beyond), reflecting the innovative research involved and offering a reliable basis for future research.
This study found that the abundance of microbes, such as Roseburia, Parabacteroides, Metascardovia, Staphylococcus, and Bacillales in the mutation group was notably higher compared to the non-mutation group. The increase in the abundance of Staphylococcus in the mutation group might increase the risk of infection in colorectal cancer patients and enhance the decomposition of intestinal substances, causing more exotoxins to be released, and resulting in abdominal pain and diarrhea in patients. Once diffused into the bloodstream through the intestinal tract, Staphylococcal exotoxins will induce systemic infection and intoxication, and damage the immune system, thus aggravating the disease in patients and promoting the proliferation and metastasis of tumors. As such, the increase in staphylococcus in the intestinal tracts of colorectal cancer patients with KRAS gene mutation could strengthen the growth capacity of the tumor compared to the non-mutation group indirectly explaining why the proliferation and metastasis capacities of the tumor in the mutation group were stronger than those in the non-mutation group and the mechanism therein.
Furthermore, the mutation group had markedly lower levels of microbes such as Clostridiales, Bacteroidetes, Lachnospiraceae, Coprococcus, and Ruminococcaceae compared to the non-mutation group. Both Bacteroidetes and Actinobacteria are colonized in the intestinal tract, and play important regulatory roles in the decomposition of intestinal substances, the balance of intestinal flora, and the intestinal immune function. The increase in the abundance of Bacteroidetes and Actinobacteria in the mutation group was conducive to regulating the intestinal environment and intensified the intestinal immune response to release related cell-active factors and tumor suppressors. This stimulates the activation of the systemic immune system and ultimately applying killing effects on colorectal cancer in the intestinal tract and the whole body. The highly positive correlation between Actinobacteria and Bacteroidetes proves that they can act synergistically to regulate the release of related immune substances in the intestinal tract, thereby inhibiting and killing tumors.
It is concluded that the prognosis of colorectal cancer patients is different between the mutation and non-mutation groups due to the differences in the composition and abundance of intestinal flora. Intestinal microbes affect the proliferation, invasion, and metastasis of tumors through the local immune system of the intestinal tract and the systemic immune system.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5622
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5622
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5622). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients were informed of this study and consented to participate in it, with the approval of the Ethics Committee in our hospital (No. K2019 08). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Gbolahan O, O’Neil B. Update on systemic therapy for colorectal cancer: biologics take sides. Transl Gastroenterol Hepatol 2019;4:9. [Crossref] [PubMed]
- Yi C, Huang Y, Yu X, et al. Clinicopathologic distribution of KRAS and BRAF mutations in a Chinese population with colorectal cancer precursor lesions. Oncotarget 2016;7:17265-74. [Crossref] [PubMed]
- Vitiello PP, Cardone C, Martini G, et al. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. J Exp Clin Cancer Res 2019;38:41. [Crossref] [PubMed]
- Tosi F, Magni E, Amatu A, et al. Effect of KRAS and BRAF Mutations on Survival of Metastatic Colorectal Cancer After Liver Resection: A Systematic Review and Meta-Analysis. Clin Colorectal Cancer 2017;16:e153-63. [Crossref] [PubMed]
- Korkmaz L, Coskun HS, Dane F, et al. Kras-mutation influences outcomes for palliative primary tumor resection in advanced colorectal cancer-a Turkish Oncology Group study. Surg Oncol 2018;27:485-9. [Crossref] [PubMed]
- Yang J, Tan Q, Fu Q, et al. Gastrointestinal microbiome and breast cancer: correlations, mechanisms and potential clinical implications. Breast Cancer 2017;24:220-8. [Crossref] [PubMed]
- Tözün N, Vardareli E. Gut Microbiome and Gastrointestinal Cancer: Les liaisons Dangereuses. J Clin Gastroenterol. 2016;50 Suppl 2, Proceedings from the 8th Probiotics, Prebiotics & New Foods for Microbiota and Human Health meeting held in Rome, Italy on September 13-15, 2015:S191-6.
- Gao R, Kong C, Huang L, et al. Mucosa-associated microbiota signature in colorectal cancer. Eur J Clin Microbiol Infect Dis 2017;36:2073-83. [Crossref] [PubMed]
- Han S, Gao J, Zhou Q, et al. Role of intestinal flora in colorectal cancer from the metabolite perspective: a systematic review. Cancer Manag Res 2018;10:199-206. [Crossref] [PubMed]
- Sideris M, Emin EI, Abdullah Z, et al. The Role of KRAS in Endometrial Cancer: A Mini-Review. Anticancer Res 2019;39:533-9. [Crossref] [PubMed]
- Ogishima J, Taguchi A, Kawata A, et al. The oncogene KRAS promotes cancer cell dissemination by stabilizing spheroid formation via the MEK pathway. BMC Cancer 2018;18:1201. [Crossref] [PubMed]
- Mao C, Wu XY, Yang ZY, et al. Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep 2015;5:8065. [Crossref] [PubMed]
- Yang X, Xu W, Tian X, et al. Diagnostic and prognostic value of KRAS mutations in circulating pancreatic ductal adenocarcinoma tumor DNA. Transl Cancer Res 2018;7:622-33. [Crossref]
- Eibl G, Rozengurt E. KRAS, YAP, and obesity in pancreatic cancer: A signaling network with multiple loops. Semin Cancer Biol 2019;54:50-62. [Crossref] [PubMed]
- Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757-65. [Crossref] [PubMed]
- Forbes S, Clements J, Dawson E, et al. COSMIC 2005. Br J Cancer 2006;94:318-22. [Crossref] [PubMed]
- Neumann J, Zeindl-Eberhart E, Kirchner T, et al. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract 2009;205:858-62. [Crossref] [PubMed]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer 2007;7:295-308. [Crossref] [PubMed]
- Yang J, Tan Q, Fu Q, et al. Gastrointestinal microbiome and breast cancer: correlations, mechanisms and potential clinical implications. Breast Cancer 2017;24:220-8. [Crossref] [PubMed]
- Barko PC, McMichael MA, Swanson KS, et al. The Gastrointestinal Microbiome: A Review. J Vet Intern Med 2018;32:9-25. [Crossref] [PubMed]
- Gopalakrishnan V, Helmink BA, Spencer CN, et al. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018;33:570-80. [Crossref] [PubMed]
- Wroblewski LE, Peek RJ, Coburn LA. The Role of the Microbiome in Gastrointestinal Cancer. Gastroenterol Clin North Am 2016;45:543-56. [Crossref] [PubMed]
- Allali I, Boukhatem N, Bouguenouch L, et al. Gut microbiome of Moroccan colorectal cancer patients. Med Microbiol Immunol 2018;207:211-25. [Crossref] [PubMed]
- Kwa M, Plottel CS, Blaser MJ, et al. The Intestinal Microbiome and Estrogen Receptor-Positive Female Breast Cancer. J Natl Cancer Inst 2016;108:djw029. [PubMed]
- Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol 2017;14:527-39. [Crossref] [PubMed]
(English Language Editor: A. Kassem)