
Diagnostic accuracy of metagenomic next-generation sequencing for active tuberculosis in clinical practice at a tertiary general hospital
Introduction
Tuberculosis (TB) is the tenth leading cause of death worldwide and the leading cause of death from a single infectious agent, ranking above HIV/AIDS. In 2017, approximately 10.0 million new TB patients, including an estimated 1.3 million HIV-negative and 0.3 million HIV-positive individuals, died of the disease worldwide. Due to its large population, China had a total of 889,000 new cases in 2017, although the incidence rate was only 63/100,000, ranking as one of the top three countries for the total TB incidence (1).
Early and accurate detection of Mycobacterium tuberculosis (MTB) is important for TB diagnosis, immediate treatment and prevention of TB transmission (2). Despite advances in diagnostics, a considerable proportion of TB cases reported to the World Health Organization (WHO) are still clinically diagnosed rather than bacteriologically confirmed. Classic diagnostic tests for TB include acid-fast bacilli (AFB) smears and mycobacterial culture. AFB tests have limitations in terms of low sensitivity and confusion with nontuberculous Mycobacteria (3). Although liquid mycobacterial culture can reduce the median turnaround time and increase sensitivity, up to 2–6 weeks is still required to provide results (4). Because microscopy fails to allow species identification, and culture-based workflows for the diagnosis of TB remain slow, technically demanding. Therefore, we need fast and accurate methods for diagnosing TB. Xpert MTB/RIF is a nucleic acid amplification test recommended by the WHO to diagnose TB that can identify MTB and rifampin (RIF) resistance simultaneously in 2 hours through probes complementary to the rpoB gene (5). Previous studies have shown the high sensitivity and specificity of the Xpert MTB/RIF assay for the detection of TB and RIF resistance; nonetheless, it has limited detection efficacy in some cases of extrapulmonary TB (EPTB) (6-8).
In general hospitals, many difficult miscellaneous diseases can affect various systems and organs, which creates difficulty in diagnosis and differential diagnosis. Rapid and accurate diagnostic techniques for pathogens are urgently needed. Metagenomic next-generation sequencing (mNGS) is an emerging rapid test for the diagnosis of different pathogens, including MTB, at the same time (9,10). However, the clinical application of mNGS in the diagnosis of TB has mostly been reported in case reports (11,12). Our study aimed to evaluate the diagnostic accuracy of mNGS in pulmonary TB (PTB) and EPTB in clinical practice. We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/atm-20-2274).
Methods
Study subjects
We retrospectively reviewed 820 samples from patients with different diseases, including infectious and non-infectious diseases, at Zhongshan Hospital in Shanghai, China, between 1 April 2017 and 31 March 2018. Specimens were subjected to regular clinical microbiological assays and mNGS testing (BGI China) in parallel. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). An ethical review application was validated by the Ethical Review Committee of Zhongshan Hospital, Fudan University, Shanghai, China (approval No. B2017-193R). Written informed consent was obtained for all patients or their legal designees.
Case definitions of TB
A case of TB refers to a patient who presents with symptoms, signs, images, microbial results or pathology suggestive of TB, where a clinician has diagnosed TB and decided to treat the patient with a full course of TB therapy (13).
Bacteriologically confirmed TB (confirmed TB) refers to a case of TB with MTB identified from a clinical specimen, either by culture or by a newer method such as Xpert MTB/RIF or TB nucleic acid PCR.
Clinical TB refers to a case of TB without the M. tuberculosis complex (MTBC) identified.
PTB refers to a case of TB involving the lung parenchyma (13).
EPTB refers to a case of TB involving organs other than the lungs, e.g., the pleura, lymph nodes, abdomen, genitourinary tract, skin, joints and bones, or meninges. Intrathoracic TB lymphadenopathy (mediastinal and/or hilar) or TB pleural effusion, without radiographic abnormalities in the lungs, constitutes a case of EPTB.
Not tuberculosis (NOT TB) refers to a case with an established alternative diagnosis.
mNGS and analysis
Different samples [sputum, bronchoalveolar lavage fluid (BALF), serous fluid, blood, tissue homogenates, etc.] were collected from patients according to standard procedures. The process of mNGS consisted of sample processing, nucleic acid extraction, library generation, and bioinformatic pipeline analysis as described previously (14). Extracted DNA was fragmented to yield 150- to 200-bp fragments, and DNA fragments were constructed through an end-repair method. Quality-qualified libraries were sequenced on the BGISEQ-100 platform. High-quality sequencing data were generated by removing low-quality and short (length <35 bp) reads, followed by computational subtraction of human host sequences mapped to a human reference genome (hg19) using Burrows-Wheeler alignment. Nonhuman sequences were mapped to classification reference databases downloaded from NCBI (
Criteria for a positive MTB mNGS result
The MTBC includes M. tuberculosis, M. canettii, M. africanum, and M. bovis in the mNGS database. TB was considered positive with at least one read mapped to the MTBC (number of sequences strictly mapped at the genus level, SMRNG ≥1). We evaluated the accuracy of mNGS for TB diagnosis.
Statistical analysis
Comparative analysis was conducted by Pearson’s test, Fisher’s exact test or the McNemar test for discrete variables where appropriate. Data analysis was performed with SPSS 22.0. P values less than 0.05 were considered significant, and all tests were two tailed.
Results
Sample and patient characteristics
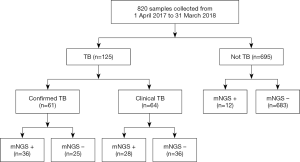
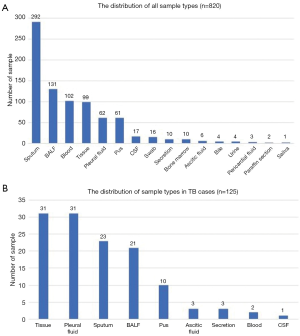
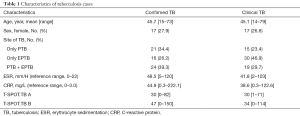
A total of 820 samples were included in the analysis, with 125 samples (15.2%) from TB cases and 695 samples (84.8%) from NOT TB cases (Figure 1). The distribution of all sample types is listed in Figure 2A, and the distribution of TB cases is shown in Figure 2B. Of all the TB cases, the number of confirmed TB cases was 61, and the number of clinical TB cases was 64. Pulmonary samples including sputum, BALF and lung tissue accounted for 58.2% of all the samples (477/820). Other common sample types included blood, pleural fluid, pus, and extrapulmonary tissue. In TB cases, extrapulmonary samples accounted for most samples [57.6% (72/125)]; specifically, 34 serous fluid samples (27.2%), 22 extrapulmonary tissue samples (17.6%), 10 pus samples (8.0%), and 6 other samples [3 blood samples, 2 secretion samples, and 1 cerebrospinal fluid (CSF) sample] were included. The pulmonary samples included 23 sputum samples (18.4%), 21 BALF samples (16.8%), and 9 lung tissue samples (7.2%). The characteristics of the TB cases are listed in Table 1.
Full table
Turnaround times for different test methods
The turnaround time for mNGS was compared with the times for AFB smears and cultures on liquid medium (the MGIT 960 system). The mNGS turnaround time in our study was 32–36 hours, which was significantly shorter than that of culture on liquid medium (median 34 days, 14–55 days). Although this turnaround time was not shorter than that of AFB smears (median 1 day, 0–1 days), AFB smears had very low sensitivity (positive for only 7 of 125 samples).
Diagnostic performance comparison of mNGS and a culture assay
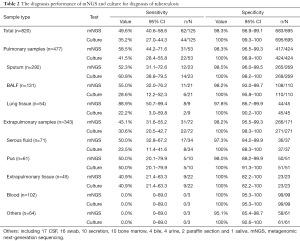
As shown in Table 2, the overall sensitivity of mNGS was 49.6% [95% confidence interval (95% CI), 40.6–58.6%], which was superior to that of the culture assay (35.2%, 95% CI, 27.0–44.3), and the overall specificity was 98.3% (95% CI, 96.9–99.1%). Although the sensitivity for pulmonary samples was much higher than that for extrapulmonary samples (58.5%, 95% CI, 44.2–71.6% versus 43.1%, 95% CI, 31.6–55.2%, respectively), no superiority was noted for the sputum between mNGS and the culture assay (52.3%, 95% CI, 31.1–72.6% versus 60.9%, 95% CI, 38.8–79.5%, respectively). Peak sensitivities were observed in lung tissues (88.9%; 95% CI, 50.7–99.4%), BALF (55.0%; 95% CI, 32.0–76.2%), and serous fluids (50.0%; 95% CI, 32.8–67.2%). The sensitivities of pus and non-lung tissues were similar for the culture assay (50.0%; 95% CI, 20.1–79.9% and 40.9%; 95% CI, 21.4–63.3%, respectively). MTB was detected in 12 samples from NOT TB cases (listed in Table 3), in which active TB at the relevant site was excluded.
Full table
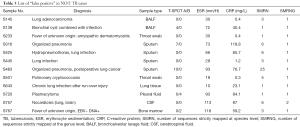
Full table
Concordance between mNGS and the culture assay
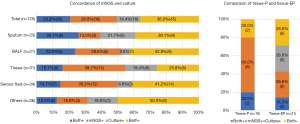
mNGS and the culture assay were both positive for 26/125 samples (20.8%) and both negative for 45/125 samples (36.0%), while 36 samples (28.8%) were positive by mNGS only and 18 samples (14.4%) were positive by the culture assay only. Positivity differed among sample types, with more mNGS positivity found in BALF, lung tissue and serous fluid (Figure 3).
Comparison of mNGS analysis between confirmed TB and clinical TB
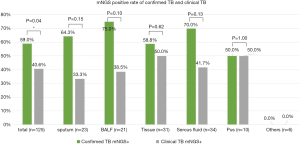
We had 61 confirmed TB cases and 64 clinical TB cases. The MTB positivity of mNGS was higher in confirmed TB than in clinical TB (59.0% versus 40.6%, respectively, P<0.05), and a similar trend was observed among various sample types, although without a significant difference. In clinical TB without conclusive aetiological evidence, mNGS substantially improved the detection of positive cases by 40.6%, performing exceptionally in tissue and pus (50.0%) (Figure 4).
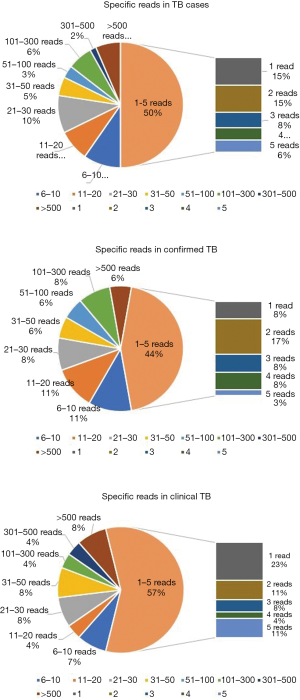
The number of unique reads of the MTBC by mNGS ranged from 1 to 3,966, and the coverage ranged from 0.01% to 99.4%. Because of the difficulty of DNA extraction, the read numbers mapped to the MTBC (SMRNG) were mostly small, and almost 50% of cases had fewer than 5 reads. No significant difference in SMRNG number was observed between confirmed TB and clinical TB (Figure 5).
Discussion
In the present study, we retrospectively reviewed TB cases with both mNGS and a culture assay performed in a pairwise manner with different sample types to illustrate the feasibility of mNGS for early diagnosis of PTB and EPTB. mNGS produced a higher sensitivity for MTB than the culture assay (49.6% and 35.2%, respectively), and a high specificity of 98.3%. The turnaround time could be shortened from 2–6 weeks to 32–36 hours and is currently reduced to almost 24 hours due to the localization of sequencing in our hospital.
Moreover, our results showed that mNGS was not advantageous with sputum, but a relatively higher sensitivity was detected for lung tissue and BALF, implying that mNGS performed better in culture-negative PTB than in culture-positive PTB, which is relatively difficult to diagnose. The overall sensitivity of mNGS in detecting MTB in EPTB appeared to be superior to that of the culture assay and not inferior to that of Xpert MTB/RIF (6-8). However, mNGS was superior to Xpert MTB/RIF in serous fluid evaluation in most studies (5). Due to the low sensitivity of the culture assay itself, the concordance between mNGS and the culture assay was not high, but mNGS could detect MTB more frequently in culture-negative specimens, which can provide more aetiological evidence in clinical practice.
MTB was not detected in 47 CSF samples, probably because of the low proportion of definite/probable tubercular meningitis. Wang et al. reported that mNGS is a good method to detect MTB in CSF samples from patients with tubercular meningitis (15). Unfortunately, in our study, the diagnostic value of mNGS in tubercular meningitis was difficult to evaluate, as we need more meningitis cases. The MTBC was not detected in blood samples. Detection of the MTBC in the blood of a recently diagnosed disseminated TB patient in our department suggested the value of peripheral blood mNGS for disseminated TB.
The MTBC was detected at the species level in 12/695 (1.5%) NOT TB cases; however, no previous data for mNGS in latent tuberculosis infection (LTBI) or other infections have been reported. Moreover, the possibility of strong positive pollution in the same batch on our NGS platform complicates explaining whether this result is a false positive or indicative of contamination.
The concordance between culture and mNGS was 56.8%, and mNGS additionally detected sequences of the MTBC in some culture-negative samples. A positive mycobacterial culture requires approximately 100 organisms per mL, and culture has reduced sensitivity in paucibacillary samples (such as serous fluid, BALF, and tissue homogenate) and samples from patients who previously received anti-TB therapy (14,16,17).
As MTB is an intracellular mycobacterial pathogen, the read number of the MTBC detected was very small, with fewer than 5 reads in half of the cases due to the difficulty of DNA extraction. Theoretically, the read number of mNGS should be positively correlated with the amount of bacteria in a specimen; while no correlation was found between MTB positivity by mNGS and bacterial confirmation, positive AFB smears, or the MTB culture period. The preprocessing procedure should be optimized to improve the application of mNGS for intracellular pathogens such as Mycobacterium, Brucella, and fungi (9). Even if only one MTBC sequence is detected, TB still needs to be highly suspected (12,14).
Some limitations exit in this study. First, this was a retrospective study with inappropriate patient selection. Second, bacterially confirmed cases were mostly evaluated by culture, and only a few cases were PCR positive or Xpert MTB/RIF positive. We could not simultaneously compare mNGS with other rapid detection methods, especially Xpert MTB/RIF, which the WHO recommends as a confirmatory test. Third, our tests were delivered to a centralized laboratory, and DNA degradation may have occurred because of inappropriate specimen delivery.
Overall, most of the cases in our study were atypical PTB or difficult-to-diagnose cases affecting various systems, and mNGS determination of MTB positivity may provide a prompt clinical diagnosis and a direction for clinical strategies. In conclusion, mNGS may be a promising technology for early diagnosis of active TB, especially sputum culture-negative PTB and tuberculous serous effusions.
Acknowledgments
The authors thank all clinicians, pathologist, and microbiologists who have assisted with the study.
Funding: This work was supported by the BGI China (precision medical funding) and Zhongshan Hospital, Clinical Research Funds of Zhongshan Hospital, Fudan University (2018ZSLC06).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-2274
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2274
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2274). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). An ethical review application was validated by the Ethical Review Committee of Zhongshan Hospital, Fudan University, Shanghai, China (approval No. B2017-193R). Written informed consent was obtained for all patients or their legal designees.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- WHO. Global tuberculosis report 2018. Available online: https://www.who.int/tb/publications/global_report/en/
- Marx FM, Yaesoubi R, Menzies NA, et al. Tuberculosis control interventions targeted to previously treated people in a high-incidence setting: a modelling study. Lancet Glob Health 2018;6:e426-e435. [Crossref] [PubMed]
- Jain D, Ghosh S, Teixeira L, et al. Pathology of pulmonary tuberculosis and non-tuberculous mycobacterial lung disease: Facts, misconceptions, and practical tips for pathologists. Semin Diagn Pathol 2017;34:518-29. [Crossref] [PubMed]
- Cruciani M, Scarparo C, Malena M, et al. Meta-analysis of BACTEC MGIT 960 and BACTEC 460 TB, with or without solid media, for detection of mycobacteria. J Clin Microbiol 2004;42:2321-5. [Crossref] [PubMed]
- World Health Organization. Automated Real-Time Nucleic Acid Amplification Technology for Rapid and Simultaneous Detection of Tuberculosis and Rifampicin Resistance: Xpert MTB/RIF Assay for the Diagnosis of Pulmonary and Extrapulmonary TB in Adults and Children: Policy Update. WHO Guidelines Approved by the Guidelines Review Committee 2013, Geneva.
- Scott LE, Beylis N, Nicol M, et al. Diagnostic accuracy of Xpert MTB/RIF for extrapulmonary tuberculosis specimens: establishing a laboratory testing algorithm for South Africa. J Clin Microbiol 2014;52:1818-23. [Crossref] [PubMed]
- Maynard-Smith L, Larke N, Peters JA, et al. Diagnostic accuracy of the Xpert MTB/RIF assay for extrapulmonary and pulmonary tuberculosis when testing non-respiratory samples: a systematic review. BMC Infect Dis 2014;14:709. [Crossref] [PubMed]
- Denkinger CM, Schumacher SG, Boehme CC, et al. Xpert MTB/RIF assay for the diagnosis of extrapulmonary tuberculosis: a systematic review and meta-analysis. Eur Respir J 2014;44:435-46. [Crossref] [PubMed]
- Simner PJ, Miller S, Carroll KC. Understanding the Promises and Hurdles of Metagenomic Next-Generation Sequencing as a Diagnostic Tool for Infectious Diseases. Clin Infect Dis 2018;66:778-88. [Crossref] [PubMed]
- Schlaberg R, Chiu CY, Miller S, et al. Validation of Metagenomic Next-Generation Sequencing Tests for Universal Pathogen Detection. Arch Pathol Lab Med 2017;141:776-86. [Crossref] [PubMed]
- Ai JW, Li Y, Cheng Q, et al. Diagnosis of local hepatic tuberculosis through next-generation sequencing: Smarter, faster and better. Clin Res Hepatol Gastroenterol 2018;42:178-81. [Crossref] [PubMed]
- Li H, Gao H, Meng H, et al. Detection of Pulmonary Infectious Pathogens From Lung Biopsy Tissues by Metagenomic Next-Generation Sequencing. Front Cell Infect Microbiol 2018;8:205. [Crossref] [PubMed]
- Chen Q, Guo X, Wang X, et al. T-SPOT.TB in Detection of Active Tuberculosis During Pregnancy: A Retrospective Study in China. Med Sci Monit 2016;22:57-60. [Crossref] [PubMed]
- Miao Q, Ma Y, Wang Q, et al. Microbiological Diagnostic Performance of Metagenomic Next-generation Sequencing When Applied to Clinical Practice. Clin Infect Dis 2018;67:S231-S240. [Crossref] [PubMed]
- Wang S, Chen Y, Wang D, et al. The Feasibility of Metagenomic Next-Generation Sequencing to Identify Pathogens Causing Tuberculous Meningitis in Cerebrospinal Fluid. Front Microbiol 2019;10:1993. [Crossref] [PubMed]
- Lewinsohn DM, Leonard MK, LoBue PA, et al. Official American Thoracic Society/Infectious Diseases Society of America/Centers for Disease Control and Prevention Clinical Practice Guidelines: Diagnosis of Tuberculosis in Adults and Children. Clin Infect Dis 2017;64:111-5. [Crossref] [PubMed]
- Fontanilla JM, Barnes A, von Reyn CF. Current diagnosis and management of peripheral tuberculous lymphadenitis. Clin Infect Dis 2011;53:555-62. [Crossref] [PubMed]


