Bioinformatic analysis and in vitro validation of a five-microRNA signature as a prognostic biomarker of hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) ranks fifth in terms of cancer mortality in the world, and h morbidity has increased each year (1). Dysregulated gene expression has been extensively demonstrated to exert a vital role in tumorigenesis and cancer progression. Growing evidence has uncovered diagnostic, prognostic, and even therapeutic targets for different types of cancers through the different gene expression profiles. Moreover, some mutant genes have recently been suggested to participate in the carcinogenesis and progression of HCC (2). However, HCC is a highly heterogeneous disease, with large variations in HCC progression and significantly complicated prognostic prediction. Thus, the urgent exploration of effective diagnostic and prognostic biomarkers is required.
MicroRNAs (miRNAs), a group of small noncoding RNA molecules, control about one-third of protein-coding genes via post-transcriptional gene regulation, thereby exerting direct or indirect effects on the majority of cellular pathways (3). The critical biological roles of miRNAs have been validated in previous research, demonstrating miRNAs influence in carcinogenesis, tumor progression, metastasis, and patient survival (4,5). A number of miRNAs have been suggested as possible diagnostic and prognostic biomarkers and promising therapeutic targets (6). Additionally, several miRNA signatures have been constructed for prognostic estimation in HCC patients (7-12). However, the common drawbacks of these studies are that they lack validation, ongoing enrollment patients, discovery of novel miRNAs, and uniformity in processing and analyzing the data. Thus, the current prognostic miRNA signatures still need to be optimized, and the identification of practical and potent miRNA signatures may play a critical role for prognostic prediction in clinical practice.
The present study was designed to establish and verify a miRNA-based classifier by using the least absolute shrinkage and selection operator (LASSO) Cox regression model. The robust miRNA expression-based signature was constructed to improve prognostic prediction of HCC by comprehensively analyzing genomic data. The model miRNA was further confirmed by in vitro analysis. We present the study in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-2509).
Methods
Patient datasets and processing
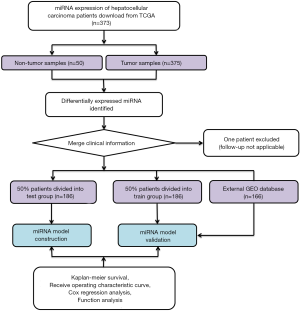
Publicly accessible raw counts of miRNA expression profiles, along with the corresponding clinical data of HCC patients, were downloaded from The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/tcga/, accessed July 2019). The data of 373 HCC patients, comprising 375 HCC cancer samples and 50 adjacent normal samples, were downloaded. The data from one patient were excluded due to a lack of follow-up information. In total, data for 372 HCC patients were randomly categorized into either a training group or a validation group at a ratio of 1 to 1 for comprehensive integrated analysis using the “caret” package. The flow chart is shown in Figure 1. This study used open-access data and so did not require additional approval from an ethics committee. Data processing was performed strictly according to the National Institutes of Health (NIH) TCGA Human Subject Protection and Data Access Policies. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data processing and screening of differentially expressed (DE) miRNAs
The raw count of miRNA from each sample was used to form the expression matrix, and was followed by the normalization of the expression matrix using “edgeR” package derived from R language. |log2 foldchange (FC)| >2.0 along with adjusted P value <0.05 was employed for the identification of DE miRNAs. Heat map and volcano plots were further performed on DE miRNAs for better differentiation between normal and HCC tumor samples.
Construction of the miRNA signature
The correlation expression of each miRNA with patient survival was calculated by univariate Cox model, where a P value <0.05 indicated statistical significance for a specific miRNA. “Survminer” package in R project was used to determine the optimal cutoff miRNA values to investigate the relationship between the miRNAs signature and the prognosis of patients. R software was further used for random number generation to equally categorize patients into the training and validation cohorts. In the training cohort LASSO was conducted using R package “glmnet” for selection of the most valuable predictive miRNAs, and was simultaneously employed to obtain shrinkage and variable selection; meanwhile, 10-time cross-validations were used to determine the optimal values of penalty parameter λ. Moreover, the risk score of the miRNA signature for each patient was calculated according to the expression of every prognostic miRNA along with its relevant coefficient, which is shown as follows: ; where Expi is the miRNA signature expression of patient i, and coefi is the LASSO coefficient of miRNA i.
Confirmation of the miRNA signature
Patients were distributed based on their risk score and survival information. HCC patients were further categorized into high- and low-risk groups with the median risk score being used as the cutoff value; this was followed by the generation of Kaplan-Meier (KM) survival curves and Cox proportional hazards regression in the two groups. The prognostic performance of miRNA risk scores was assessed by a time-dependent receive operating characteristic (ROC) curve via the comparison of sensitivity and specificity. Multivariate Cox regression analysis was subsequently conducted to confirm whether the predictive performance of miRNA risk score was independent of other clinicopathological parameters. Additionally, the predictive performance of the miRNA model was verified in the validation and entire groups. A two-sided P value <0.05 suggested statistical significance, and R language (version 3.6.0; R Foundation) was used for all analyses.
miRNA signature validation using the OncomiR database and the GEO dataset
To further test the proposed miRNA-derived prognostic value, we used the OncomiR online database (http://www.oncomir.org/) to cross-validate the reliability of the model. Using OncomiR, we analyzed our miRNA signature for HCC survival outcome. The user-defined signature in predicting overall survival (OS) was demonstrated through KM curves and log-rank tests. The Gene Expression Omnibus (GEO) GSE31384 dataset was used as an external validation cohort (n=166). Patients were also classified according to the median risk score threshold as high- or low-risk, and KM survival curves were plotted.
Functional analysis
StarBase (http://starbase.sysu.edu.cn/) was employed in predicting mRNAs specific to miRNAs. Using StarBase, we searched the interactions of miRNA-targets using multiple target-predicting programs (PITA, RNA22, miRmap, DIANA-microT, miRanda, PicTar, and TargetScan). Then, protein-coding genes with significance were selected to analyze the functional enrichment using Metascape (http://metascape.org) (13). Gene set enrichment analysis (GSEA), using JAVA program (http://software.broadinstitute.org/gsea/index.jsp), was employed for assessing the possible mechanisms). The number of random sample permutations was set at 1,000, and a P value <0.05 was set as the significance threshold.
In vitro analysis
Cells from one human normal hepatocyte cell line, MIHA (RRID:CVCL_SA11), and two hepatocyte cell lines, HepG2 (RRID:CVCL_0027) and HCCLM3 (RRID:CVCL_6832) were provided by the Shanghai Cell Bank of the Chinese Academy of Sciences, and cultivated within Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Invitrogen). Cells were then cultivated in the absence of antibiotics at 37 °C in an atmosphere of 5% CO2 and 99% relative humidity. They were then transfected with miR-137 mimic (miR-137) or the negative control scramble miR (miR-NC) according to the manufacturer’s instructions (purchased from Guangzhou RIBOBIO, China). The total cellular or tissue RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) in accordance with manufacture protocols. The 2−ΔΔCt approach was utilized to normalize target RNA expression. HepG2 (1×103 cells per well) and HCCLM3 (1×103 cells per well) cells were seeded in 96-well plates. Cell viability was measured with the Cell Counting Kit-8 (CCK-8, Dojindo). Transwell assay was carried out to determine cell migratory capacity. In the migration assay, transduced cells were inoculated into the 24-well plates. After 24 h, the cells were serum-starved overnight and followed by trypsin digestion. Later, 200 µL serum-free medium containing 5×104 cells was placed into the upper chamber, while 700 µL culture medium supplemented with 10% FBS was placed into the lower chamber to grow for 24 h at 37 °C. The subsequent migratory cells were subjected to 4% paraformaldehyde fixation and 0.5% crystal violet staining, and the number of cells stained was calculated under the microscope. In addition, HCC cell lines were cultivated within the culture dish, scratched with a 10-µL pipette tip, and cultured in FBS-free DMEM. Finally, the inverted microscope (OLYMPUS IX73) was used to observe the migratory cells at 24 h post injury.
Statistical analysis
LASSO regression was conducted to identify potential miRNAs. Survival outcomes were compared according to the KM method using the survival package in R. Time-dependent ROC curves were constructed using the R software package “survival ROC”. Cox regression was conducted to verify the relationship of miRNA risk score prediction with other clinical parameters. Comparisons between groups were performed with Student’s t-test and one-way ANOVA. A two-sided P value <0.05 was defined as statistically significant. All statistical analyses were performed using R (https://www.r-project.org) software.
Results
Clinical features
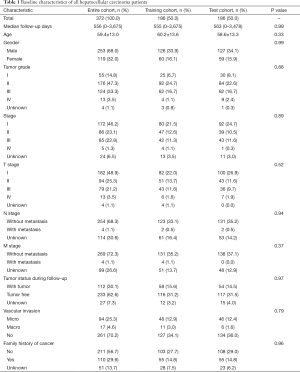
The baseline characteristics of all HCC patients are shown in Table 1. All samples were randomly categorized into two groups: a training cohort (n=186) and a testing cohort (n=186). The clinical features of patients in the training and testing cohorts were required to be similar. The expression profiles and clinical data from the training and testing cohorts are shown in files (https://cdn.amegroups.cn/static/application/15aac4fbcd5b505fe70f39214ebb9de2/ATM-20-2509-1.xlsx and https://cdn.amegroups.cn/static/application/e178bf8ebc5c5590f86bad217abc42ee/ATM-20-2509-2.xlsx), respectively.
Full table
Identification of DE miRNAs
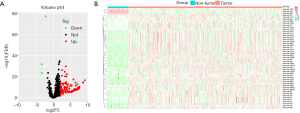
In total, 127 DE miRNAs were identified between tumor samples and non-tumor samples (https://cdn.amegroups.cn/static/application/008ed35cca4ff295542c5299b6e9d29d/ATM-20-2509-3.xlsx), including 124 upregulated miRNAs and 3 downregulated miRNAs. A heat map was used to display the top DE miRNAs (Figure 2A), and a volcano plot was used to show miRNAs with significant differences between cancer and normal samples (Figure 2B).
Construction of the miRNA signature
Univariate Cox regression analysis revealed a significant association between 16 DE miRNAs and survival. Using “survminer” package, we determined the optimal cutoff miRNAs values, and the 16 corresponding survival curves are shown in Figure 3. LASSO regression was further used for validation of variables in the training cohort (Figure 4A,B), giving rise to five miRNAs, including has-mir137, hs-mir-105-2, has-mir-548f-1, has-mir-9-3, and has-mir-561. The forest plot of the correlation between each miRNA and survival is shown in Figure 4C. The prognostic risk score was imputed as follows: (0.6078 × has-mir-105-2) + (0.8071 × has-mir-9-3) + (0.8529 × has-mir-137) + (0.6551 × has-mir-548f-1) + (0.7866 × has-mir-561).
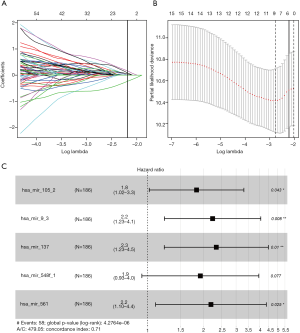
Confirmation of the miRNA signature
A risk score was calculated for each patient, and the patients were divided into high- or low-risk groups based on the median cutoff value. Risk score and survival time distributions in the training group are shown in Figure 5A,B, respectively. KM curves for high- and low-risk groups are displayed in Figure 5C, and show poorer OS in patients with high risk scores (P=5.682e-6). The ROC curve was further plotted for assessing the prognostic capacity of miRNAs biomarkers. The area under the curve (AUC) of the miRNA biomarker prognostic model was 0.726 (Figure 5D). The AUC of miRNA model was higher than that of every single miRNA or TNM stage (Figure S1). In the univariate Cox proportional hazards regression model, the hazard ratio (HR) of risk score was 2.979 [95% CI (confidence interval): 1.661–5.341] (Figure 5E), which was consistent with the outcomes from the multivariate Cox proportional hazards regression model (HR = 3.285, 95% CI: 1.737–6.213) after adjustment of the clinical covariate (Figure 5F).
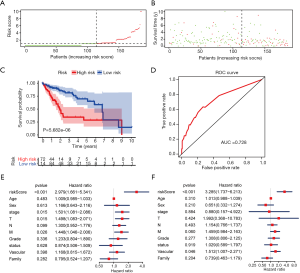
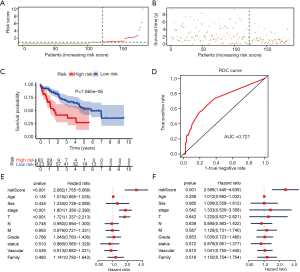
miRNA model in the validation cohort, OncomiR, and GEO
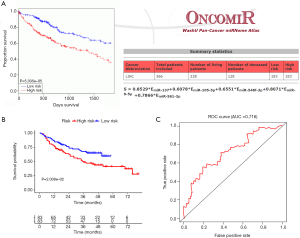
To determine if the proposed miRNA model exerted similar prognostic significance in other groups, we applied the same formula in the validation cohort. The miRNA risk score and survival time distributions in the validation group are shown in Figure 6A,B, respectively. KM curves in the high- and low-risk groups are displayed in Figure 6C. Patients with a low-risk score had a survival benefit in the validation group (P=7.865e-6). The AUC for the validation group was 0.721 (Figure 6D). The miRNA model AUC was also higher than every single miRNA or TNM stage AUC (Figure S1). Similar to the training cohort, univariate and multivariate analyses of the validation cohort demonstrated the presently established miRNA model as an independent prognostic indicator (Figure 6E,F). A similar pattern was seen in OncomiR. Patients with a high risk score had poor OS (P=5.035e-5, Figure 7A). To confirm the external validity, the model was applied in the external GEO data. Figure 7B shows the high-risk group had a significantly shorter survival than the low-risk group in the GSE31384 cohorts (P=0.020). ROC curve analysis showed that the risk signature prognosis prediction could attain an AUC value of 0.716 (Figure 7C).
Functional analysis of the prognostic miRNA model
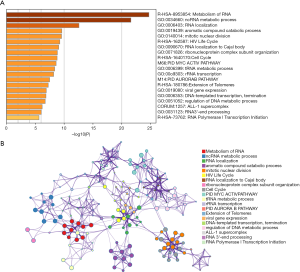
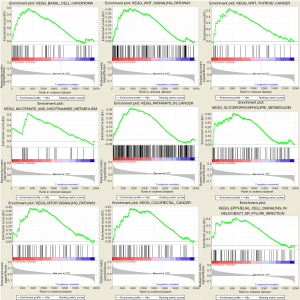
For in-depth investigation of the possible biological effects of these identified miRNAs, miRNA-related mRNAs were filtered out. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted on these selected mRNAs using Metascape. Specifically, terms having the P value of <0.01 and ≥3 enriched genes were regarded as significant. The top 20 pathways screened from GO and KEGG analyses are shown in Figure 8. GSEA was further conducted to illustrate the biological roles of the miRNA model, and indicated that the high-scoring miRNA model showed significant enrichment in Wnt signaling pathways, mTOR signaling pathways, and several cancer pathways (Figure 9).
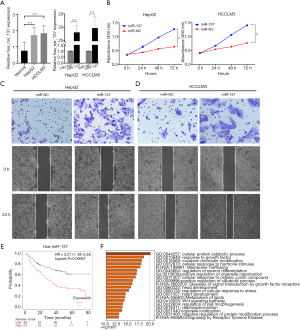
miR-137 contributed to HCC progression and its target genes prediction
As miR-137 expression was the most upregulated of the five prognostic miRNAs in HCC, we performed in vitro experiments to further investigate the role of miR-137. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) showed that the miR-137 expression was high in HCC cell lines, and significantly increased following transfection (Figure 10A). We then performed the CCK-8 assay to detect the effect of miR-137 overexpression on cell proliferation. After miR-137 overexpression, HepG2 and HCCLM3 cell proliferation significantly increased compared with the miR-NC groups (Figure 10B, P<0.001). Transwell was conducted in order to determine the miR-137 role in the migratory capacity of HCC cells. Relative to controls, ectopic miR-137 expression within HepG2 (Figure 10C) and HCCLM3 (Figure 10D) cell lines resulted in the markedly upregulated migratory capacities of cells. A wound healing assay revealed that miR-137 overexpression significantly accelerated wound healing in HepG2 and HCCLM3 cells. A high miR-137 level correlated with poorer survival according to KM Plotter online tools (Figure 10E). The target genes of miR-137 were predicted, enrichment-analyzed, and are visualized in Figure 10F. Catenin-beta-interacting protein 1 (CTNNBIP1), a negative regulator of the Wnt signaling pathway, was predicted to be the target gene of miR-137 (https://cdn.amegroups.cn/static/application/aeb3ecf6ed3e8488be77e16af0f66be8/ATM-20-2509-4.xlsx). The above results suggested that miR-137 promotes HCC progression.
Discussion
To date, only a small number of miRNA-based models on HCC prognosis have been established, with common drawbacks including small study populations, inconsistent approaches to processing and analyzing data, and a lack of validation. To overcome these limitations, we conducted the current research in HCC patients to predict their survival using miRNA signatures. DE miRNAs that significantly correlated with survival were comprehensively screened by applying biostatistical methods and univariate Cox analysis. Five miRNAs were subsequently filtered out by using LASSO regression on the miRNA data in TCGA. Significant miRNAs were further incorporated for the establishment of prognostic models. KM curves, time-dependent ROC curves, and Cox regression analysis, confirmed the five-miRNA signature to be an independent prognostic predictor in HCC. Further validation was carried out in the validation group with OncomiR and GEO being used to confirm the reliability of our results. Functional analyses including GO, KEGG, and GSEA were conducted to demonstrate the biological role of the miRNA model in HCC progression. Finally, we conducted in vitro assays to evaluate the promotive role of has-mir-137 in HCC cell proliferation and migration. Shi et al. constructed an 11-miRNA expression signature in HCC using datasets published prior to 2013 from the GEO, Web of Science, and ArrayExpress databases (8). However, as the data were published many years ago an and data homogeneity was as an issue, the data should be interpreted cautiously. Wang et al. identified a miRNA profile in hepatitis B virus-induced HCC (14), while Fu et al. performed an analysis specifically for resectable HCC (11). Following this, Liao et al. (10) and Bai et al. (12) identified potential miRNA biomarkers for prognostic prediction in HCC, while Lin et al. (7) and Han et al. (9) proposed miRNA signatures for vascular invasion to predict survival in HCC. However, the abovementioned studies only used conventional Cox regression rather than LASSO regression for model construction and thus lacked further validation.
Our study design was superior to other research concerning the prognostic significance of miRNAs in HCC in a number of ways. To begin, all HCC patients in the TCGA group were enrolled for analysis, with a considerable sample size. Second, unlike the conventional stepwise regression in previous research, the accuracy of bioinformatics analysis was enhanced by LASSO penalized regression, which is capable of simultaneously analyzing all independent parameters and identifying the most influential ones (15). After introduction of a penalty following a regularization path, the coefficients of variables with less influence become zero (16). Thus, the LASSO approach provides significantly higher accuracy compared with the conventional multivariate Cox stepwise regression model, particularly in cases of extremely large datasets, like genomics (17). Third, to ensure the reliability of the results, we produced the miRNA signature in both the training group and the validated model. Finally, we conducted an in vitro study to clarify the functional role of miR-137.
Although a number of miRNAs have been reported to be associated with tumorigenesis and tumor progression, the functions of most miRNAs remain largely undefined. Anwar et al. (18) found that aberrant hypermethylation and concomitantly decreased expression of intergenic miRNA genes were commonly detected in HCC; one of these genes, hsa-mir-9-3 (50%), was also identified by Potapova to be a novel target of abnormal DNA methylation in human HCC (19). Additionally, miR-137 might function as a prognostic biomarker in HCC population, which is also a possible target for eliminating liver cancer stem cells (20-22). Previous studies have also reported that miR-137 has either cancer cell-promoting or -suppressing capabilities (23). miR-137 was reported to be upregulated and to promote invasion in breast, bladder, non-small cell lung cancer (24-26), and more controversially, in liver cancer. Some research has found miR-137 to be downregulated in liver cancer, inhibiting tumor progression (22,27,28). However, other recent studies found miR-137 to have the opposite effect. Sakabe et al. found that patients with high miR-137 expression had worse survival prognosis, more frequent invasion of vessels and the bile duct, and high alpha-fetoprotein levels (20). Furthermore, Wei et al. showed that miR-137 upregulation in HCC contributed to poor survival, and could bind to Afamin, promoting invasion and metastasis of HCC cell lines (29). Our data also implicated miR-137 in promoting invasive growth of HCC cell lines, while its elevated expression was associated with worse survival of HCC patients. Another study performed miR-137-predicted target gene analysis and found that Wnt/β-catenin inhibitory protein, CTNNBIP1, could inhibit liver tumor progression and invasion (30). The combinatory effect of miR‐137 and target gene, CTNNBIP1, on the regulation of liver cancer remains to be explored. In addition to hsa-mir-9-3 and miR-137, our results, for the first time, indicate that three other miRNAs (has-mir-105-2, has-mir-548f-1, and hsa-mir-561) may be possible prognostic predictors for HCC; further in-depth studies are warranted to confirm the present findings and to examine their molecular features.
Pathway enrichment analysis showed that these miRNAs are likely to impact the carcinogenesis and progression of HCC by pathways in a fashion according to their respective biological functions in HCC. Moreover, a portion of the enriched pathways reported in literature are involved in the carcinogenesis and progression of HCC. Although the biological roles of the miRNAs contributing to HCC remain unclear, pathway enrichment analysis showed that these miRNAs are likely to impact the carcinogenesis and progression of HCC via the mTOR (31,32) or Wnt signaling pathway (33-37).
A few limitations in this study should also be addressed. First, the prognostic effects of miRNA signature in HCCs were not validated by further functional assays, but were deduced from online datasets using bioinformatics approaches. Second, while the exploration of circulatory plasmid/serum miRNAs in the circulating biofluids as potential biomarkers for cancer diagnosis and prognosis has attracted increasing attention, whether or not the proposed miRNAs can be cancer-specific and reliably detected in the blood remains unclear. Thus, further verification of the outcomes of this research is still required. Third, in vitro results need further verification, such as miRNA inhibitor experiment.
In summary, using comprehensive bioinformatics analysis and the genetic profiles and clinical features of a selected cohort, we demonstrated that a novel five-miRNA signature could be an independent prognostic biomarker for HCC. Further studies are warranted to validate the possible functions and mechanisms of these miRNAs in HCC progression.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81801804).
Footnote
Reporting Checklist: The authors present the study in accordance with the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-2509
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form(available at: http://dx.doi.org/10.21037/atm-20-2509). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen QF, Huang T, Shen L, et al. Predictive value of a nomogram for hepatocellular carcinoma with brain metastasis at initial diagnosis: A population-based study. PLoS One 2019;14:e0209293. [Crossref] [PubMed]
- Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. [Crossref] [PubMed]
- Song JL, Nigam P, Tektas SS, et al. microRNA regulation of Wnt signaling pathways in development and disease. Cell Signal 2015;27:1380-91. [Crossref] [PubMed]
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014;20:460-9. [Crossref] [PubMed]
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014;9:287-314. [Crossref] [PubMed]
- Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology 2008;47:897-907. [Crossref] [PubMed]
- Lin Z, Cai YJ, Chen RC, et al. A microRNA expression profile for vascular invasion can predict overall survival in hepatocellular carcinoma. Clin Chim Acta 2017;469:171-9. [Crossref] [PubMed]
- Shi KQ, Lin Z, Chen XJ, et al. Hepatocellular carcinoma associated microRNA expression signature: integrated bioinformatics analysis, experimental validation and clinical significance. Oncotarget 2015;6:25093-108. [Crossref] [PubMed]
- Han B, Zheng Y, Wang L, et al. A novel microRNA signature predicts vascular invasion in hepatocellular carcinoma. J Cell Physiol 2019;234:20859-68. [Crossref] [PubMed]
- Liao X, Zhu G, Huang R, et al. Identification of potential prognostic microRNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cancer Manag Res 2018;10:787-803. [Crossref] [PubMed]
- Fu Q, Yang F, Xiang T, et al. A novel microRNA signature predicts survival in liver hepatocellular carcinoma after hepatectomy. Sci Rep 2018;8:7933. [Crossref] [PubMed]
- Bai F, Zhou H, Ma M, et al. A novel RNA sequencing-based miRNA signature predicts with recurrence and outcome of hepatocellular carcinoma. Mol Oncol 2018;12:1125-37. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Wang G, Dong F, Xu Z, et al. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer 2017;17:805. [Crossref] [PubMed]
- Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010;33:1-22. [Crossref] [PubMed]
- Gao J, Kwan PW, Shi D. Sparse kernel learning with LASSO and Bayesian inference algorithm. Neural Netw 2010;23:257-64. [Crossref] [PubMed]
- McNeish DM. Using Lasso for Predictor Selection and to Assuage Overfitting: A Method Long Overlooked in Behavioral Sciences. Multivariate Behav Res 2015;50:471-84. [Crossref] [PubMed]
- Anwar SL, Albat C, Krech T, et al. Concordant hypermethylation of intergenic microRNA genes in human hepatocellular carcinoma as new diagnostic and prognostic marker. Int J Cancer 2013;133:660-70. [Crossref] [PubMed]
- Potapova A, Albat C, Hasemeier B, et al. Systematic cross-validation of 454 sequencing and pyrosequencing for the exact quantification of DNA methylation patterns with single CpG resolution. BMC Biotechnol 2011;11:6. [Crossref] [PubMed]
- Sakabe T, Azumi J, Umekita Y, et al. Prognostic relevance of miR-137 in patients with hepatocellular carcinoma. Liver Int 2017;37:271-9. [Crossref] [PubMed]
- Gao M, Liu L, Li S, et al. Inhibition of cell proliferation and metastasis of human hepatocellular carcinoma by miR-137 is regulated by CDC42. Oncol Rep 2015;34:2523-32. [Crossref] [PubMed]
- Liu LL, Lu SX, Li M, et al. FoxD3-regulated microRNA-137 suppresses tumour growth and metastasis in human hepatocellular carcinoma by targeting AKT2. Oncotarget 2014;5:5113-24. [Crossref] [PubMed]
- Mahmoudi E, Cairns MJ. MiR-137: an important player in neural development and neoplastic transformation. Mol Psychiatry 2017;22:44-55. [Crossref] [PubMed]
- Ying X, Sun Y, He P. MicroRNA-137 inhibits BMP7 to enhance the epithelial-mesenchymal transition of breast cancer cells. Oncotarget 2017;8:18348-58. [Crossref] [PubMed]
- Xiu Y, Liu Z, Xia S, et al. MicroRNA-137 upregulation increases bladder cancer cell proliferation and invasion by targeting PAQR3. PLoS One 2014;9:e109734. [Crossref] [PubMed]
- Chang TH, Tsai MF, Gow CH, et al. Upregulation of microRNA-137 expression by Slug promotes tumor invasion and metastasis of non-small cell lung cancer cells through suppression of TFAP2C. Cancer Lett 2017;402:190-202. [Crossref] [PubMed]
- Zhu M, Li M, Wang T, et al. MicroRNA-137 represses FBI-1 to inhibit proliferation and in vitro invasion and migration of hepatocellular carcinoma cells. Tumour Biol 2016;37:13995-4008. [Crossref] [PubMed]
- Wu DC, Zhang MF, Su SG, et al. HEY2, a target of miR-137, indicates poor outcomes and promotes cell proliferation and migration in hepatocellular carcinoma. Oncotarget 2016;7:38052-63. [Crossref] [PubMed]
- Wei Q, Zhao L, Jiang L, et al. Prognostic relevance of miR-137 and its liver microenvironment regulatory target gene AFM in hepatocellular carcinoma. J Cell Physiol 2019;234:11888-99. [Crossref] [PubMed]
- Fu X, Zhu X, Qin F, et al. Linc00210 drives Wnt/beta-catenin signaling activation and liver tumor progression through CTNNBIP1-dependent manner. Mol Cancer 2018;17:73. [Crossref] [PubMed]
- Matter MS, Decaens T, Andersen JB, et al. Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. J Hepatol 2014;60:855-65. [Crossref] [PubMed]
- Malakar P, Shilo A, Mogilevsky A, et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res 2017;77:1155-67. [Crossref] [PubMed]
- Dai B, Ma Y, Yang T, et al. Synergistic effect of berberine and HMQ1611 impairs cell proliferation and migration by regulating Wnt signaling pathway in hepatocellular carcinoma. Phytother Res 2019;33:745-55. [Crossref] [PubMed]
- Li B, Cao Y, Meng G, et al. Targeting glutaminase 1 attenuates stemness properties in hepatocellular carcinoma by increasing reactive oxygen species and suppressing Wnt/beta-catenin pathway. EBioMedicine 2019;39:239-54. [Crossref] [PubMed]
- Hu P, Ke C, Guo X, et al. Both glypican-3/Wnt/beta-catenin signaling pathway and autophagy contributed to the inhibitory effect of curcumin on hepatocellular carcinoma. Dig Liver Dis 2019;51:120-6. [Crossref] [PubMed]
- Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/beta-catenin pathway. Arch Biochem Biophys 2019;661:196-202. [Crossref] [PubMed]
- Huynh H, Ong R, Goh KY, et al. Sorafenib/MEK inhibitor combination inhibits tumor growth and the Wnt/betacatenin pathway in xenograft models of hepatocellular carcinoma. Int J Oncol 2019;54:1123-33. [PubMed]
(English Language Editors: J. Brown and J. Gray)