
High resolution computed tomography for the diagnosis of 2019 novel coronavirus (2019-nCoV) pneumonia: a study from multiple medical centers in western China
Introduction
Since December 2019, a succession of cases of pneumonia with unknown causes has been observed in Wuhan, Hubei Province, China. In subsequent months, this kind of pneumonia has also been reported in China and other countries (1). The causative agent of this pneumonia has been confirmed to be a novel coronavirus (2019-nCoV) from the same lineage as the coronavirus that causes severe acute respiratory syndrome (SARS), but with significantly different genetic characteristics from that of SARS-CoV and Middle East respiratory syndrome (MERS-CoV) (1-3). On January 7, 2020, the 2019 novel coronavirus (SARS-Cov2, with the disease now officially named COVID-19 by the World Health Organization) was identified as the causative agent, based on virus typing (1,4). The coronavirus enters cells by binding to the angiotensin converting enzyme 2 (ACE2) receptor which is found on the apical surface of differentiated ciliated respiratory epithelia (5-7). This cell type is particularly dense in airway epithelial cells, hence the severity of COVID-19 disease in lungs and distal airways. Due to the similar clinical manifestations and computed tomography (CT) features of viral pneumonia, it is easy to misdiagnose the disease in its early phase. A series of studies have demonstrated that the coronavirus not only exhibits animal-to-human transmission, but also human-to-human transmission (8-12). Respiratory droplet transmission and contact transmission, which is the main route of transmission, cause rapid transmission with a higher incidence rate. For this type of communicable infection which can spread rapidly, accurate and early diagnosis is crucial for appropriate treatment, and can help save lives and control the epidemic. At present, the diagnosis of COVID-19 infection mainly depends on the detection of viral nucleic acid through reverse transcription polymerase chain reaction (RTPCR) of sputum, throat swabs, lower respiratory tract secretions, and other specimens (13). However, due to sampling errors, the sensitivity for diagnosis of the disease is unstable, and the nucleic acid test is time-consuming. Furthermore, the reports about 2019-nCoV pneumonia mainly refer to epidemiologic characteristics, clinical manifestations, laboratory findings, and treatment outcomes of the patients.
High-resolution computed tomography (HRCT) plays a central and important role in the evaluation and diagnosis of focal lung disease, which can detect lesions and indicate if they are virus-derived infections (14). However, to the best of our knowledge, few existing studies have evaluated the diagnostic values of HRCT combined with epidemiological history from multiple medical centers in western China (low risk area), and compared the diagnostic accuracy of HRCT features with at least two time viral nucleic acid examinations. Therefore, the purpose of this study was to evaluate HRCT diagnostic values for 2019-nCoV pneumonia combined with epidemiological history and provide imaging evidence for clinical diagnosis and appropriate treatment. We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5731).
Methods
Patient population
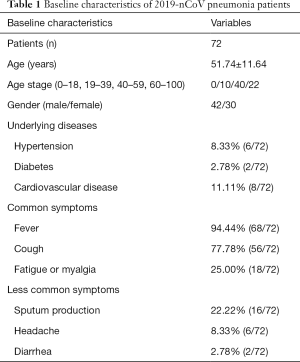
This retrospective, multicenter study was conducted according to principles of the Declaration of Helsinki (as revised in 2013) and approved by the ethics commissions of the Third Hospital of Mianyang (MYTH2020-19). The need for written informed consent from the participants was waived. The patient’s personal data have been secured. The study included 72 consecutive patients (30 women and 42 men; mean age 51.74 years; age range: 23–78 years) confirmed to have 2019-nCoV infection, who had undergone HRCT of the chest at our five hospitals(the Third Hospital of Mianyang, Suining Central Hospital, the Affiliated Hospital of North Sichuan Medical College, the First People’s Hospital of Neijiang, and the Affiliated Yongchuan Hospital of Chongqing Medical University). The patients were categorized as per the severity of the disease into early stage (n=36), progressive stage (n=32), and severe stage (n=4) using the guidelines for Imaging Diagnosis of Novel Coronavirus (first edition, 2020) (15). Patient demographic characteristics are shown in Table 1.
Full table
HRCT imaging protocol and image analysis
High-resolution CT scans were obtained on a variety of scanners, including Aquilion ONE TSX-301 (Toshiba, Japan), SOMATOM Definition AS+ (Siemens, Germany), Brilliance iCT (Philips, Netherlands) and GE Light speed VCT (GE, America). Images were acquired using following parameters: 2–4 mm section thickness; 20–40 mAs; 120–150 kV. Multi-plane reconstruction (MPR) technology was used. The HRCT features were analyzed by 2 radiologists with 10 or more years of experience in reading pulmonary CT. Both of them were blinded to the results of detection of viral nucleic acid.
Epidemiological criteria and detection of viral nucleic acid
The epidemiological criteria to define a suspected case were as follows: history of travel to Wuhan city or direct contact with patients from Wuhan who had symptoms of lower respiratory illness (e.g., cough, breathing difficulties), within 14 days before illness onset; history of fever or symptoms of lower respiratory illness and close contact with patients confirmed with 2019-nCoV infection (16). Respiratory specimens were tested for 2019-nCoV pneumonia using real-time fluorescence polymerase chain reaction (RT-PCR), with nasal and pharyngeal swabs, sputum, bronchial aspirates, and bronchoalveolar lavage fluid.
Statistical analyses
Statistical analysis was performed with SPSS (Version 23.0, IBM Corp., USA). Categorical variables are expressed as number percentages. The diagnosis accuracy between HRCT features combined with epidemiological history and the first detection of viral nucleic acid was compared using χ2 test or Fisher’s exact test. A P value <0.05 was considered statistically significant.
Results
Baseline characteristics
By February 7th, 2020, 72 patients who had undergone HRCT of the chest at our five institutions were confirmed to have 2019-nCoV infection; 10 (13.89%) of the 2019-nCoV-infected patients were aged 19–39 years, 40 (55.56%) were aged 40–59 years, and 22 (30.56%) were aged over 60 years; no children or adolescents were infected. The median age of the patients was 51.74 years, with age ranging from 23 to 78 years. The clinical and demographic characteristics of the patients are summarized in Table 1.
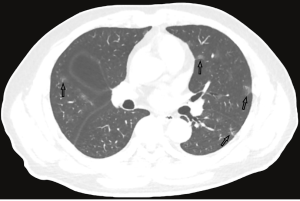
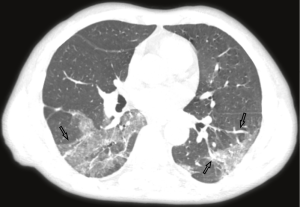
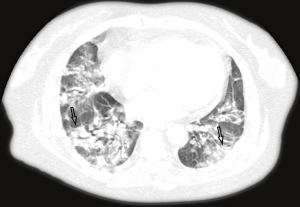
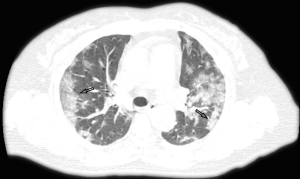
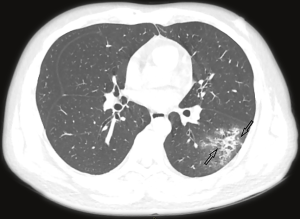
The majority of the infected patients were men [42 (58.33%) of 72]; less than a quarter had underlying diseases [16 (22.22%)], including hypertension [6 (8.33%)], diabetes [2 (2.78%)], and cardiovascular disease [8 (11.11%)]. Common symptoms at onset of 2019-nCoV pneumonia were fever [68 (94.44%) of 72 patients], cough [56 (77.78%)], and fatigue or myalgia [18 (25.00%) of 72]; less common symptoms were sputum production [16 (22.22%)], headache [6 (8.33%) of 72], and diarrhea [2 (2.78%) of 72]. Laboratory studies of the patients on admission showed leucopenia (white blood cell count less than 4×109/L; 20 (27.78%) of 72 patients),lymphopenia [lymphocyte count <1.0×109 /L; 44 (61.11%) patients], and elevated C-reactive protein [C-reactive protein count >10 mg/L; 36 (50.00%) patients]. Out of these 72 patients, 36 were diagnosed as early stage, 32 patients as progressive stage, and 4 patient as severe stage. Figures 1-3 show the representative HRCT appearance on images of three cases confirmed as early stage, progressive stage, and severe stage, respectively.
CT imaging findings
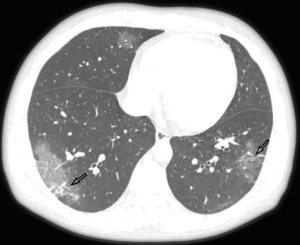
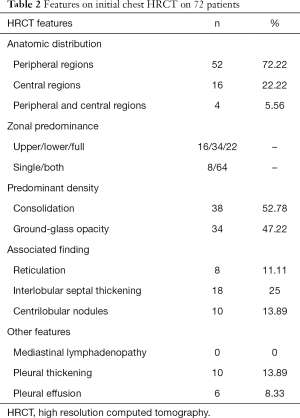
All patients had lung parenchymal abnormalities on HRCT scans (Figures 1-6), which were mostly multifocal in both lungs and asymmetric in all patients. They were mostly in the peripheral or subpleural lung regions in 52 patients (72.22%) (Figure 1,2), in the central lung regions in 16 patients (22.22%), and in both lungs with “white lung” manifestations in 4 patients (5.56%) (Figure 3). The CT image features mainly included consolidation, ground-glass opacity (GGO), interlobular septal thickening, centrilobular nodules, and reticulation. Subpleural multifocal consolidation was the predominant abnormality in 38 patients (52.78%) (Figure 4). GGO was seen in 34 patients (47.22%) (Figure 5). Interlobular septal thickening was found in 18 (25.00%) patients, 8 of whom had only generally mild thickening with no zonal predominance. Reticulation was seen in 8 patients (11.11%), and was mild and randomly distributed (Figure 6). In addition, both lungs of 28 patients had 2 or 3 CT imaging features.
The overall extent of abnormalities was less than 25% of the lung parenchyma in 38 patients, 25–50% in 30, 50–75% in 4, and no one had more than 75% abnormalities. CT manifestations were divided into early stage, progressive stage, severe stage, and absorption stage according to the Guidelines for Imaging Diagnosis of Novel Coronavirus (first edition, 2020). Table 2 summarizes the high-resolution CT findings.
Full table
Comparison of the diagnosis accuracy between HRCT features and RT-PCR
All of the cases were proven 2019-nCoV pneumonia patients by RT-PCR. The detection rate of the first nucleic acid test was 88.89%; meanwhile, the detection rate was 91.67% for the combination of two tests, 97.22%, for the combination of three tests, and 100% for the combination of four tests. The diagnostic accuracy of HRCT features combined with epidemiological history was not significantly different from the diagnosis by the detection of viral nucleic acid (all P>0.05). A comparison of the diagnostic accuracy of HRCT features combined with epidemiological history with that of the first detection of viral nucleic acid is shown in Table 3.

Full table
Discussion
Since the outbreak of the new coronavirus pneumonia (2019-nCov) epidemic, the number of deaths caused by the new coronavirus pneumonia has been increasing with rise of the number of daily diagnoses (17). Therefore, how to diagnose the disease accurately and quickly is crucial for appropriate treatment, and thus can help save lives and control the epidemic.
The results of the present study demonstrated that the diagnostic accuracy of HRCT features combined with epidemiological history is similar to detection via viral nucleic acid. While viral nucleic acid testing is considered the gold standard for the diagnosis of 2019-nCoV pneumonia (13), there remain a few disadvantages: first, the nucleic acid test is time-consuming, and if it is negative, repeated sampling may be necessary, which would prolong the diagnostic time and increase chance of the further spread of the virus; second, although the specificity of the nucleic acid test is high, the false-negative rate is still appreciable. These shortcomings make coping with a large number of close contacts difficult.
For chest HRCT examination, although it lacks the specificity for viral pneumonia and may appear as a different disease with the same imaging features, typical HRCT features combined with epidemiological history have a sensitivity similar to that of viral nucleic acid. It is acknowledged that chest CT cannot replace nucleic acids to confirm the diagnosis of new coronavirus pneumonia. However, in the face of conditions where nucleic acid testing is not feasible in the epidemic area and a large number of suspected cases cannot be diagnosed, chest HRCT examination combined with epidemiological history can be used as a powerful supplementary method. It can perform leak detection and make up for defects, so as to find more suspected cases and close contacts in the early phase, allowing patients to be isolated, observed, and treated as soon as possible.
According to the Guidelines for Imaging Diagnosis of Novel Coronavirus (first edition; 2020) (15), our study cases were classified as early stage (n=36), progressive stage (n=32), and severe stage (n=4). In early stage imaging findings, lesions were mostly distributed in the peripheral or subpleural lung and generally do not involve the whole lung segment. At the same time, the lesions were always multifocal in both lungs and rarely single focus, commonly manifesting as GGOs, with or without interlobular septal thickening, and rarely with a small and limited consolidation range. When the disease was in the progressive stage, the distribution of lesions was increased and involved multiple lobes, mostly located in the subpleural region of the lungs. Some of the lesions were enlarged with increased density, and were irregular, fan-shaped, or wedge-shaped, with unclear boundaries, showing bilateral asymmetry. If treatment was delayed or treated inappropriately, diffuse lesions could be seen in both lungs, with a few “white lung” manifestations. The range of lesions increased by 50% within 48 hours, and the lesions were mainly consolidated, with GGOs, air bronchogram, and multiple cable-like shadows.
Our study found that 10 (13.89%) of the 2019-nCoV-infected patients were aged 19–39 years, 40 (55.56%) were aged 40–59 years, and 22 (30.56%) were aged over 60 years. However, no children or adolescents were infected. We acknowledge that these results might be implausible, but may be explained by the fact that the children have few opportunities to go out and have less chance of contacting the source of infection; therefore, the probability of infection in children could be low.
This study indeed has several limitations. Foremost, the sample size in our study was relatively small and lacking cases of children or adolescents. Further studies with increased patient population are recommended. Secondly, we did not count false-positive cases of CT imaging, as these cases were not included in this study, which could lead to potential inclusion bias and weaken our findings. Thirdly, the patients underwent the CT scans with different machine type, due to the multiple centers in the study. The heterogeneity of the CT data might affect the results of the study. Finally, we did not review the follow-up CT imaging, Long-term follow-up is required in future to provide dynamic CT evaluation for pulmonary lesions and to obtain the data of long term pulmonary function changes.
In conclusion, the HRCT features of 2019-nCoV pneumonia are sufficiently characteristic to assess scope and type of lesions, which, when combined with epidemiological history, yield high clinical value for the diagnosis of 2019-nCoV pneumonia.
Acknowledgments
Funding: This work was supported by Emergency Scientific Research Project of the 2019-nCoV Pneumonia in Mianyang City Sichuan Province (No. 2020YJKY004).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5731
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5731
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5731). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective, multicenter study was conducted according to principles of the Declaration of Helsinki (as revised in 2013) and approved by the ethics commissions of the Third Hospital of Mianyang (MYTH2020-19). The need for written informed consent from the participants was waived. The patient’s personal data have been secured.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 2020;382:727-33. [Crossref] [PubMed]
- Paraskevis D, Kostaki EG, Magiorkinis G, et al. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol 2020;79:104212. [Crossref] [PubMed]
- Chen Y, Liu Q, Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol 2020;92:418-23. [Crossref] [PubMed]
- Jia HP, Look DC, Shi L, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol 2005;79:14614-21. [Crossref] [PubMed]
- Ren X, Glende J, Al-Falah M, et al. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J Gen Virol 2006;87:1691-5. [Crossref] [PubMed]
- To KF, Lo AW. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2). J Pathol 2004;203:740-3. [Crossref] [PubMed]
- Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 2018;23:130-7. [Crossref] [PubMed]
- Li Q, Guan X, Wu P, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med 2020;382:1199-207. [Crossref] [PubMed]
- Carlos WG, Dela Cruz CS, Cao B, et al. Novel Wuhan (2019-nCoV) Coronavirus. Am J Respir Crit Care Med 2020;201:7-8. [Crossref] [PubMed]
- Wu P, Hao X, Lau EHY, et al. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Euro Surveill 2020;25:2000044. [Crossref] [PubMed]
- Gralinski LE, Menachery VD. Return of the Coronavirus: 2019-nCoV. Viruses 2020;12:135. [Crossref] [PubMed]
- Nishiura H, Jung SM, Linton NM, et al. The Extent of Transmission of Novel Coronavirus in Wuhan, China, 2020. J Clin Med 2020;9:330. [Crossref] [PubMed]
- Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020;25:2000045. [Crossref] [PubMed]
- Chung M, Bernheim A, Mei X, et al. CT Imaging Features of 2019 Novel Coronavirus (2019-nCoV). Radiology 2020;295:202-7. [Crossref] [PubMed]
- Guidelines for Imaging Diagnosis of novel Coronavirus. Available online: https://www.cmtopdr.com/post/detail/c1c25295-9a92-48fe-a58d-c97e334173f7
- Guidelines for Diagnosis and Treatment of novel Coronavirus (5th edtion). Available online: http://www.chinacdc.cn/jkzt/crb/zl/szkb_11803/jszl_11815/202002/t20200205_212256.html
- Guan WJ, Ni ZY, Hu Y, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med 2020;382:1708-20. [Crossref] [PubMed]
(English Language Editor: J. Gray)


