
Unique genomic features and prognostic value of COSMIC mutational signature 4 in lung adenocarcinoma and lung squamous cell carcinoma
Introduction
Lung cancer causes the highest number of both recent cases (2.09 million) and deaths (1.76 million) among all cancers worldwide in 2018 (1). In China, lung cancer is also the most commonly diagnosed cancer (18.1%) and the most common cause of cancer-related deaths (30.9%) (2). Non-small cell lung carcinoma (NSCLC) accounts for 80% to 85% of lung cancers. Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the major histological types of NSCLC, and together make up approximately 70% of all cases of lung cancers (3-5). Cigarette smoking is the primary contributor to LUADs and LUSCs, but a substantial portion of LUADs are from non-smokers (6). LUADs of non-smokers are more common in females and occur more frequently in younger people than other types of lung cancer (7). Although some environmental factors, including second-hand smoking and occupational exposure to carcinogens, are correlated with LUADs of non-smokers, the cause of tumorigenesis in non-smoking LUADs remains unknown (6).
Genetic alterations in LUADs and LUSCs are primarily distinct. The fundamental oncogenic alterations in LUADs, including point mutations in EGFR and KRAS and gene fusions comprising ALK, RET, and ROS1, are rarely found in LUSCs (8,9). TP53 mutation is dominant in LUSCs, reaching nearly 90% cases (8). Apart from the genomic variations, epigenetic modifications, including hypermethylation of CDK13, RUNX3 and APC, and hypomethylation of CDKN2A and MGMT, were magnificent in LUADs but not LUSCs (10). These differences in either genome or epigenome explain that drugs targeted for LUADs are unsuitable for LUSCs in most cases.
Genetic comparison between non-smokers and smokers has been performed multiple times in LUADs and LUSCs. It has been determined that the amplification frequency of FGF19, FGF3, FGF4, and CCND1 was five-times higher in smokers than non-smokers in LUSCs (11). Point mutations in EGFR are more frequent in non-smokers than in smokers of LUADs (9). Although the above discoveries highlighted the difference of oncogene alterations between LUADs and LUSCs, the mutational signatures and the driver genes related to them are still unknown.
In this study, we aim to characterize the genomic landscape of Chinese LUADs and LUSCs grouped by mutational signatures on the one hand and to explore the mutational process in them, however. We found that mutational signature 4 is the most prevalent mutational signature in LUSC patients and the second prevalent in LUAD patients. We found that signature 4 is a prognostic factor in both LUADs and LUSCs, although the finding in LUSCs is less robust because of the limited patient number. Different gene mutation features, including single nucleotide variation of CSMD3, LRP1B, TP53, SYNE1, and SLIT2 and copy number variation of FGF4 and FGF19, displayed in both LUADs and LUSCs when they are further grouped by mutational signature 4 status. We present this article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5952).
Methods
Patients and samples
Specimens from 493 Chinese lung cancer patients, including 401 LUADs and 92 LUSCs, were collected from 2015 to 2019. Patients were selected mainly based on the following three rules. First, they were clinically diagnosed as primary LUAD or LUSC. Second, collected tissue/biopsy samples passed quality check and gave quality sequencing data. Third, clinical and follow-up information are available for analysis. All of them have signed informed consent. From each patient, both tumor tissue and matched normal blood samples were collected. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Tongji Hospital (No. K-W-2020-014) and informed consent was taken from all the patients.
Whole exome sequencing (WES) analysis
WES and analysis were performed at the Genomics Laboratory of GenomicCare Biotechnology (Shanghai, China). For thawed soft tissue or blood, DNA was extracted using the Maxwell RSC Blood DNA Kit (cat# AS1400, Promega, Madison, WI, USA) on a Maxwell RSC system (cat# AS4500, Promega). For formalin-fixed, paraffin-embedded (FFPE) tissue, DNA was extracted using the MagMAX FFPE DNA/RNA Ultra Kit (cat# A31881, ThermoFisher, Waltham, MA, USA) on a KingFisher Flex system (ThermoFisher). The extracted DNA was sheared using a Covaris L220 sonicator. Then, the exome DNA was captured using the SureSelect Human All Exon V7 kit (cat# 5991-9039 EN, Agilent). After that, it was prepared for the library using the SureSelectXT Low Input Target Enrichment and Library Preparation system (cat# G9703-90000, Agilent, Santa Clara, CA, USA), and sequenced on an Illumina NovaSeq-6000 sequencer (Illumina, San Diego, CA, USA) to generate 150×150 bp paired-end reads. Image analysis and base calling were done using Illumina onboard RT3 software (Illumina). After removing adapters and low-quality reads, the reads were aligned to NCBI human genome reference assembly hg19 using the Burrows-Wheeler Aligner alignment algorithm and further processed using the Genome Analysis Toolkit (GATK, version 3.5), including the GATK Realigner Target Creator to identify regions that needed to be realigned.
Bioinformatic analysis
The aligned sequences of tumor tissue and whole blood samples were compared to call somatic mutations. Somatic single-nucleotide variants (SNV), Indel, and copy number variation (CNV) were determined using the MuTect/ANNOVAR/dbNSFP31, VarscanIndel, and CNVnator software, respectively, as reported in (12). The mutational signature classification was from COSMIC Mutational Signature (version 2 – March 2015), which were generated from studies performed by others (13-15). Tumor mutation burden (TMB) was defined by the number of somatic nonsynonymous mutations using a previously described method for WES data (16).
Statistical analysis
Statistical analysis and data visualization were processed by R (version 4.0.0) and GraphPad Prism 8. The package MAFtools was implemented in R.
Results
Characteristics of LUADs and LUSCs
Tumor tissue and matched normal blood samples were collected from 401 LUAD and 92 LUSC patients from 2015 to 2019. The tumor content was assessed by independent pathologists to confirm that it was above 20%, the minimum value required by our bioinformatic analysis pipeline to give an accurate calling of somatic mutations. The clinical characteristics of the patients, excluding the missing information, are shown in Table 1. The median age is similar between LUAD and LUSC (59 vs. 61 years). The median survival time is also similar between LUADs and LUSCs either (20 vs. 20.5 months) (Table 1).
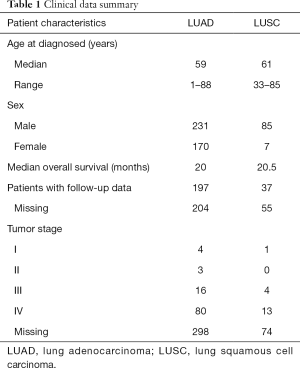
Full table
Mutational signature distribution in LUADs and LUSCs
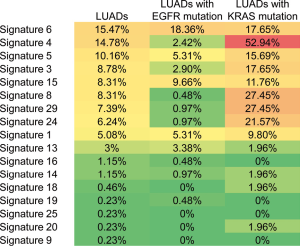
During oncogenesis, mutagenic forces leave mutational “scares” characteristic to each of them as specific changes of individual nucleotides and their combinations on the genome. Alexandrov et al. define these characteristic changes as “mutational signatures” (17-19). Some mutational signatures have been studied extensively and proven valuable in guiding cancer treatment and prevention (20,21). To learn the mutational signatures in the LUAD and LUSC patients, we analyzed their WES data and extracted mutational signatures according to the COSMIC database (version 2), which includes 30 defined signatures. In the LUAD patients, the ten most frequent signatures are 6, 4, 5, 3, 8, 15, 29, 24, 1, and 13 ordered in frequency from high to low and ranged from ~16% to 5% of patients (Figure 1). The list of top 10 signatures in the LUSC patients is almost identical, but their occurrence rates and order differ from the LUAD patients (Figure 1). A higher part of LUSC patients has identifiable mutational signatures than LUAD patients.
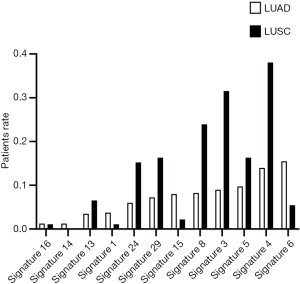
The most notable signature among the top 10 lists in both LUADs and LUSCs is signature 4, suggested associating with smoking (17). Its occurrence frequency is the highest in LUSC (~38%) and the second highest in LUAD (~16%). However, not necessarily all smokers with LUAD and LUSC have the scare of signature 4 in their genome (6). Because of the limitation of clinical records, we only know the smoking history of 25 LUAD patients. Among those we know their smoking history, signature 4 was discovered in 7 (28%) patients. These findings showed that at least a portion of smokers with LUADs (72% of the known cases) do not harbor signature 4, consistent with the above statistics from literature. The findings favored our analysis in that about one-third of LUAD smokers displayed minor to no contribution of signature 4 (7).
Signature 3 is another notable signature as it is associated with the failure of DNA double-strand break repair by homologous recombination (17). The most prevalent mutational signature in LUADs was signature 6, associated with defective DNA mismatch repair. Both signatures 3 and 6 favor cancer progression and the accumulation of mutations. It is interesting to find that the rates of signatures 6 and 15 in LUSC are lower than those in LUAD, while the rates of other significant signatures in LUSC are always higher than in LUAD. Signature 6 and 15 are characterized predominantly by C>T transition and share similar mutational features, which are associated with many insignificant (shorter than 3 bp) insertions and deletions at mono/polynucleotide repeats (17). Although 79 environmental agents have been evaluated for their effects on the induction of mutational signatures (22), the mutagens of signature 6 and 15 are still unexplained. An understanding of the cause of signatures 6 and 15 would contribute to the explanation of this phenomenon.
The prognosis of patients with different signatures in LUADs and LUSCs
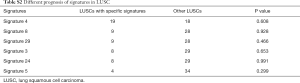
To test the possibility of using the mutational signatures in lung cancer as prognostic predictors, we evaluated the survival of patients with or without specific top-ranked signatures, one at a time, in LUADs and LUSCs. In LUADs, patients with signatures 4, 8, 29 and 24, respectively, showed a worse survival compared with the patients without the indicated individual signatures (P=0.0031, P=0.013, P<0.001 and P=0.002, respectively) (Figure 2A and Table S1). It has been reported that signature 4 exhibits transcriptional strand bias for C>A mutations and is also associated with CC>AA dinucleotide substitutions. Signatures 8, 29, and 24 all exhibit strand bias for C>A substitutions. Signature 8 and 29 are associated with CC>AA double nucleotide substitutions (13,17). Therefore, there is some similarity between these four mutational signatures. Almost all LUAD patients with signature 8, signature 29, or signature 24 were segregated compared to those with signature 4 (Figure S1), suggesting these four signatures are clustered together. In LUSC, signature 4 showed a correlation with worse survival but did not reach a significant threshold, probably because of the limited sample size in this group (Figure 2B). Only 38 LUSC patients have effective survival data. The survival of LUAD and LUSC patients cannot be distinguished according to the other mutational signatures (Tables S1,S2).
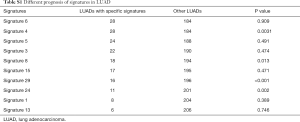
Full table

Full table
Genome-wide mutational process in LUADs and LUSCs
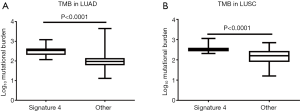
Besides a worse survival prognosis, the TMB is much higher in patients with signature 4 than those without in both LUAD and LUSC (P<0.0001, Figure 3). Mutation load has been confirmed as a predictor for immune checkpoint inhibition (ICI) therapy in NSCLC (23,24). The above results suggested that patients with signature 4 could receive help from ICI treatment.
EGFR and TP53 are the top two mutated genes and present in almost half of LUAD patients (48%, Figure 4A), which is consistent with a previous report that studied 128 Chinese LUADs (25). Interestingly, there are no other common genes, except KRAS, on the top mutated gene lists in our study and the other report. None of these genes mutated in over 15% of patients in both studies; that may explain the discrepancy (<15%). It may also be because of the divergence of mutational processes since the first oncogenesis induced by mutations on EGFR or TP53.
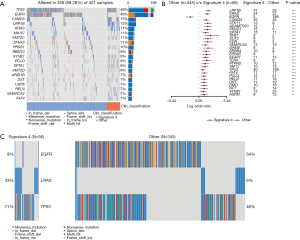
When LUAD patients were grouped according to their signature 4 status, correlated to survival, a striking difference was observed. Thirty-six genes were mutated differentially between the two groups (P<0.01) (Figure 4B,C). Among them, only one gene, EGFR, was enriched in patients without signature 4 (54%), and the rest 35 genes were more enriched in patients with signature 4. KRAS was mutated in signature 4 LUADs (42%), versus 7% in LUADs without signature 4, showing that KRAS mutation was strongly related to signature 4. Also, a little higher TP53 mutation was observed in signature 4 LUADs (69% vs. 45% LUAD without signature 4). To follow the same logic of this finding, we evaluated the association of different signatures with EGFR or KRAS mutations, respectively. We found an apparent enrichment of signature 4 in LUADs with KRAS mutation (52.94%), but there was no biased distribution of other signatures in LUADs with or without EGFR mutation (Figure S2). Besides EGFR and TP53, the most common oncogenes in LUAD, LRP1B was the next most significant gene with different distribution between LUAD patients with and without signature 4. Previous research on somatic mutations in LUAD has revealed that LRP1B was among the top mutated gene in that study (26). Moreover, Xiao et al. compared the genetic alterations in LUADs with and without the chronic obstructive pulmonary disease (COPD). They found a higher prevalence of LRP1B among the LUADs with COPD (27). Cigarette smoking is one of the major risk factors correlated with COPD (28,29). To some extent, our results showed a potential link between signature 4 and smoking. Also, CSMD3 mutation is rarely observed in LUADs in Caucasians (26,30). In our cohort, CSMD3 was the third most mutated gene in LUADs and had a much higher prevalence in signature 4 LUADs. These are one of the primary differentially mutated genes between Asians and Caucasians. Liu et al. showed that loss of CSMD3 results in increased proliferation of airway epithelial cells in NSCLC (31). Furthermore, CSMD3 was found to be the most significant single gene mutation resistant to etoposide (32). Besides these top mutated genes, mutations in other genes, including STK11, ADAMTS20, PKHD1, SPTA1, and TERT, also support that signature 4 is a preferred mutation scar in LUADs.
We also analyzed the sites of mutation in three well-known genes in LUAD, EGFR, KRAS, and TP53. In EGFR, the most frequent mutation is L858M/Q/R in the amino acid sequence, consistent with previous findings (33). However, we found this type of mutation only in patients without signature 4 (Figure S3A). In KRAS, the missense and hot spot mutations, including G12V/A/D/C/S and G13D/C, were similar in all patients irrespective of their status of signature 4 (Figure S3B). The DNA binding domain of TP53 is a mutation hot spot. The most frequent one is Y220C in patients without signature 4 (Figure S3C). There were insignificant differences in mutational site distribution in KRAS. However, some differences were observed in EGFR and TP53 between LUADs with/without signature 4.
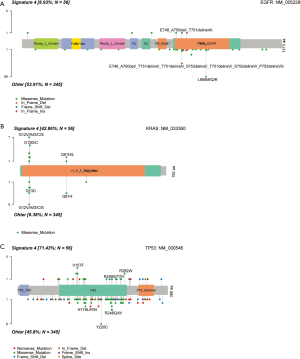
The most dominant mutated gene in LUSC is TP53 at a frequency of 76%, far higher than the subsequent frequent gene, LRP1B, at 27% (Figure 5A). Seventeen genes were found to have different mutation frequencies between LUSC patients with and without signature 4 (Figure 5B). All of them have a higher mutation rate in signature 4 LUSCs, and this is consistent with a higher mutation load in this patient group. The mutation frequency of TP53 reached 94% in LUSC patients with signature 4 (Figure 5C). There was no mutation of DPYD, ITGA10, and KIAA1549 in LUSCs without signature 4, but they reached 14% in signature 4 LUSCs. The enrichment of gene mutations of CSMD3 and LRP1B was observed in signature 4 LUSCs. These findings intrigued us to investigate whether there were more common mutated genes between these two groups (LUAD and LUSC) marked by signature 4, although LUAD and LUSC have different molecular characteristics. Indeed, six genes were present both in LUADs and LUSCs with signature 4, namely CSMD3, LRP1B, TP53, SYNE1, SLIT2, and ERBB4. Other research found that tumor suppressor SYNE1 was frequently methylated in lung cancers, and that is not associated with age at diagnosis, smoking status, or stage of lung cancer (34,35). SLIT2 can suppress lung cancer progression, but its low expression or mutation was correlated with pathological stage and reduced survival in lung cancer patients (36,37). Different types of mutations in ERBB4 have been found in NSCLC, for example, point mutation and gene fusion (38,39). These mutated genes were appointed as “Signature 4 genes” from now on.
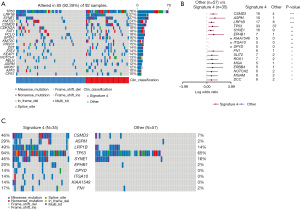
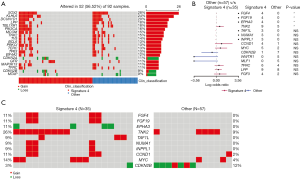
In addition to base substitutions and indels, we also analyzed CNVs in LUADs/LUSCs. Deletions at CDKN2A, MTAP, CDKN2B, and TUSC3 loci and amplification at EGFR, TERT, NKX2-1, SDHA, and NFKB1A loci were found in LUADs (Figure 6A). The difference in copy number gain of HIST2H3A is the most significant between LUAD patients with and without signature 4 (Figure 6B). No patients in the non-signature 4 group had no CNV of ERBB2, while the gain of ERBB2 was found in 7% signature 4 LUADs (Figure 6C). The gene gained most in LUAD patients without signature 4 was EGFR, at 21% versus 5% in LUAD patients with signature 4. Interestingly, an amplification region having CCND1, FGF3, FGF4, and FGF19 was found eight times more frequent in LUADs with signature 4 than without. Moreover, the amplification of EGFR and ERBB2 and the region having the above four genes occurred exclusively in LUADs with signature 4, indicating a highly variable chromosome structure even in the same type of patients.
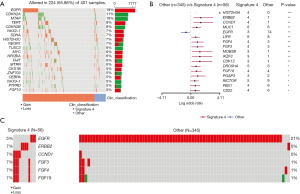
In LUSC patients, SOX2, KLHL6, DCUN1D1, LPP, and TBL1XR1 were found to be the top amplified genes, while top deleted genes were like what we found in LUADs, including CDKN2A, CDKN2B and MTAP (Figure 7A). There is significant amplification of three genes (FGF4, FGF19, and TNK2) and significant deletion of one gene (EPHA3) in signature 4 LUSCs (Figure 7B). Among them, FGF4 and FGF19 were also observed in signature 4 LUADs, so their CNV is related to signature 4 in NSCLC. Although it was not statistically significant, more patients with signature 4 had amplification of genes, including TAF1L, NUMA1, INPPL1, CCND1, and MYC (Figure 7C). These novel genes, in terms of CNV, were found to be associated with signature 4 LUSCs.
Discussion
Alexandrov and other researchers brought us a deep understanding of mutational signatures in human cancer (13,17-19,40,41), but detailed information on genetic variation in some signatures is still missing. Related studies in Asian populations are even fewer. A considerable body of research has shown the difference in gene mutation patterns within the same types of cancer between Asians and Caucasians (25,42). In this article, we analyzed the pattern of mutational signatures in Chinese LUADs and LUSCs and explored genetic features associated with the patients and mutational signatures.
We discovered that patients with and without signature 4 have different survival prospects. Signature 4 was enriched in LUAD, LUSC, small cell lung carcinoma, head and neck squamous, and liver cancers, and most of them are attributed to tobacco smoking (17,41). Although signature 4 was associated with smoking, signature 6 was found to be the most enriched in LUADs in our analysis. According to previous analyses, signature 6 is strongly correlated with dMMR and MSI-H (17). However, the MSI score of LUADs with signature 6 was less than 1%, a meager MSI score. This observation is consistent with published data (43). Additionally, signature 6 was reported to be the most common in colorectal and uterine cancers (17). The distinct characteristics of the same signature were displayed in different cancers.
Unlike MSI, high TMB is a common phenomenon in NSCLC and has been recognized as a predictor for immune checkpoint inhibition (ICI) therapy in NSCLC and melanoma (44,45). Our data showed that there was a positive correlation between signature 4 and high TMB. These results suggest the mutational signature could also be a patient stratification parameter like TMB. For instance, their dominant mutational signatures clustered esophageal adenocarcinoma patients into three subgroups, and the patients were benefited from group-specific therapies (20).
All the differentially mutated genes in LUADs with signature 4 were more enriched, except for EGFR. As we mentioned above, LRP1B mutation was one of the genes that we found to be tightly correlated with signature 4. STK11 mutation was identified to contribute to tumor heterogeneity in KRAS-mutant LUAD (46). Furthermore, it has been reported STK11 alteration is the primary cause of resistance to PD-1 blockade in KRAS-mutant murine LUAD, suggesting co-occurrence of KRAS mutation and STK11 mutation may be an adverse prognostic factor for ICI therapy (47). CSMD3, LRP1B, TP53, SYNE1, SLIT2, and ERBB4 were found to be mutated both in LUADs and LUSCs with signature 4, so may represent the genes associated with mutational signature 4 in NSCLC in general. Except for TP53 and LRP1B, the other four genes were not often reported in other lung cancer genomics studies. In addition to base substitution, gene fusion (partnered with RNF43) was observed in SYNE1 in lung cancer (48). SYNE1 is also methylated in lung cancer cell lines and LUAD (35). Wildtype SLIT2 inhibits lung cancer invasion in a ROBO-dependent manner (49). However, its attenuation was correlated with poor prognosis during lung cancer progression by deregulating beta-catenin and E-cadherin (37). Like other ERBB family members, mutant ERBB4 appears to confer “oncogene” property (50). Some studies have confirmed activating driver mutations of ERBB4 in NSCLC, which could promote the proliferation of lung cancer cells (38,51).
CNV of FGF4 and FGF19 can be regarded as “Signature 4 CNV” as they were detected in patients with signature 4 in both LUAD and LUSC. FGFs/FGFRs play a vital role in tumorigenesis by promoting cell proliferation and metastasis (52,53). FGF19 is a hormone-like enterokinase released postprandially that has recently emerged as a potential therapeutic agent for metabolic disorders, including diabetes and obesity (54). It has been found that CCND1, another well-established proto-oncogene, and FGF19 are related on a chromosomal level; further, they both are amplified in lung cancer patients (11,55). Recent data showed that the FGF19-FGFR4 signaling axis might be a key driver in certain forms of hepatocellular carcinoma (HCC), raising a strong interest in therapeutic inhibition of the pathway in this disease setting (56,57). The specificity of these amplifications suggested some typical mechanisms of tumor driving mutations in LUAD and LUSC patients with signature 4.
Conclusions
In summary, this study brought an overview of mutational signature distribution in LUADs and LUSCs of Chinese patients. Specific mutational genes and CNV were found to be associated with signature 4, associated with patient prognosis. These genetic characteristics would help individuals understand the molecular mechanism of NSCLC profoundly and supply more options for diagnosis and drug development in NSCLC.
Acknowledgments
The authors want to thank all the patients that took part in this study.
Funding: Chun Luo is supported by the Scientific Research Program of Shanghai Science and Technology Commission 2018 (grant number: 18411962500) and Shanghai Natural Science Foundation 2018 (18ZR1434500). Xiuyu Cai is supported by CSCO-Hengrui Oncology Research Fund (Y-HR2018-001).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5952
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5952
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5952). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Tongji Hospital (No. K-W-2020-014) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39:22. [Crossref] [PubMed]
- Chiba R, Morikawa N, Sera K, et al. Elemental and mutational analysis of lung tissue in lung adenocarcinoma patients. Transl Lung Cancer Res 2019;8:S224-34. [Crossref] [PubMed]
- Samriddhi S, Sajid K, Trygve OT, et al. Genetics and Epigenetics of Lung Cancer: Mechanisms and Future Perspectives. Curr Cancer Ther Rev 2013;9:97-110. [Crossref]
- Yoneyama R, Saji H, Kato Y, et al. Clinicopathological characteristics and treatment strategies for young lung cancer patients. Ann Transl Med 2019;7:100. [Crossref] [PubMed]
- Zhou F, Zhou C. Lung cancer in never smokers-the East Asian experience. Transl Lung Cancer Res 2018;7:450-463. [Crossref] [PubMed]
- Lee JJ, Park S, Park H, et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell 2019;177:1842-57.e21. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Huang T, Li J, Zhang C, et al. Distinguishing Lung Adenocarcinoma from Lung Squamous Cell Carcinoma by Two Hypomethylated and Three Hypermethylated Genes: A Meta-Analysis. PLoS One 2016;11:e0149088. [Crossref] [PubMed]
- Tan Q, Li F, Wang G, et al. Identification of FGF19 as a prognostic marker and potential driver gene of lung squamous cell carcinomas in Chinese smoking patients. Oncotarget 2016;7:18394-402. [Crossref] [PubMed]
- Zang YS, Dai C, Xu X, et al. Comprehensive analysis of potential immunotherapy genomic biomarkers in 1000 Chinese patients with cancer. Cancer Med 2019;8:4699-708. [Crossref] [PubMed]
- Alexandrov LB, Jones PH, Wedge DC, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402-7. [Crossref] [PubMed]
- Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47-54. [Crossref] [PubMed]
- India Project Team of the International Cancer Genome Consortium. Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat Commun 2013;4:2873. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246-59. [Crossref] [PubMed]
- Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev 2014;24:52-60. [Crossref] [PubMed]
- Secrier M, Li X, de Silva N, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet 2016;48:1131-41. [Crossref] [PubMed]
- Watkins JA, Irshad S, Grigoriadis A, et al. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res 2014;16:211. [Crossref] [PubMed]
- Kucab JE, Zou X, Morganella S, et al. A Compendium of Mutational Signatures of Environmental Agents. Cell 2019;177:821-36.e16. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Zhang XC, Wang J, Shao GG, et al. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun 2019;10:1772. [Crossref] [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [Crossref] [PubMed]
- Xiao D, Li F, Pan H, et al. Integrative analysis of genomic sequencing data reveals higher prevalence of LRP1B mutations in lung adenocarcinoma patients with COPD. Sci Rep 2017;7:2121. [Crossref] [PubMed]
- Miravitlles M, de la Roza C, Naberan K, et al. Use of spirometry and patterns of prescribing in COPD in primary care. Respir Med 2007;101:1753-60. [Crossref] [PubMed]
- Woodruff PG, Barr RG, Bleecker E, et al. Clinical Significance of Symptoms in Smokers with Preserved Pulmonary Function. N Engl J Med 2016;374:1811-21. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Liu P, Morrison C, Wang L, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis 2012;33:1270-6. [Crossref] [PubMed]
- Qiu Z, Lin A, Li K, et al. A novel mutation panel for predicting etoposide resistance in small-cell lung cancer. Drug Des Devel Ther 2019;13:2021-41. [Crossref] [PubMed]
- Pilotto S, Rossi A, Vavalà T, et al. Outcomes of First-Generation EGFR-TKIs Against Non-Small-Cell Lung Cancer Harboring Uncommon EGFR Mutations: A Post Hoc Analysis of the BE-POSITIVE Study. Clin Lung Cancer 2018;19:93-104. [Crossref] [PubMed]
- Tessema M, Belinsky SA. Mining the epigenome for methylated genes in lung cancer. Proc Am Thorac Soc 2008;5:806-10. [Crossref] [PubMed]
- Tessema M, Willink R, Do K, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res 2008;68:1707-14. [Crossref] [PubMed]
- Lin YY, Yang CH, Sheu GT, et al. A novel exon 15-deleted, splicing variant of Slit2 shows potential for growth inhibition in addition to invasion inhibition in lung cancer. Cancer 2011;117:3404-15. [Crossref] [PubMed]
- Tseng RC, Lee SH, Hsu HS, et al. SLIT2 attenuation during lung cancer progression deregulates beta-catenin and E-cadherin and associates with poor prognosis. Cancer Res 2010;70:543-51. [Crossref] [PubMed]
- Kurppa KJ, Denessiouk K, Johnson MS, et al. Activating ERBB4 mutations in non-small cell lung cancer. Oncogene 2016;35:1283-91. [Crossref] [PubMed]
- Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res 2014;20:3087-93. [Crossref] [PubMed]
- Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94-101. [Crossref] [PubMed]
- Pfeifer GP, Denissenko MF, Olivier M, et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 2002;21:7435-51. [Crossref] [PubMed]
- Zhang G, Wang Y, Chen B, et al. Characterization of frequently mutated cancer genes in Chinese breast tumors: a comparison of Chinese and TCGA cohorts. Ann Transl Med 2019;7:179. [Crossref] [PubMed]
- Takamochi K, Takahashi F, Suehara Y, et al. DNA mismatch repair deficiency in surgically resected lung adenocarcinoma: Microsatellite instability analysis using the Promega panel. Lung Cancer 2017;110:26-31. [Crossref] [PubMed]
- Johnson DB, Frampton GM, Rioth MJ, et al. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol Res 2016;4:959-67. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Schabath MB, Welsh EA, Fulp WJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2016;35:3209-16. [Crossref] [PubMed]
- Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov 2018;8:822-35. [Crossref] [PubMed]
- Li Y, Xiao X, Bossé Y, et al. Genetic interaction analysis among oncogenesis-related genes revealed novel genes and networks in lung cancer development. Oncotarget 2019;10:1760-74. [Crossref] [PubMed]
- Kong R, Yi F, Wen P, et al. Myo9b is a key player in SLIT/ROBO-mediated lung tumor suppression. J Clin Invest 2015;125:4407-20. [Crossref] [PubMed]
- Prickett TD, Agrawal NS, Wei X, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet 2009;41:1127-32. [Crossref] [PubMed]
- Starr A, Greif J, Vexler A, et al. ErbB4 increases the proliferation potential of human lung cancer cells and its blockage can be used as a target for anti-cancer therapy. Int J Cancer 2006;119:269-74. [Crossref] [PubMed]
- Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer 2017;17:318-32. [Crossref] [PubMed]
- Porta R, Borea R, Coelho A, et al. FGFR a promising druggable target in cancer: Molecular biology and new drugs. Crit Rev Oncol Hematol 2017;113:256-67. [Crossref] [PubMed]
- Stanley S, Buettner C. FGF19: How gut talks to brain to keep your sugar down. Mol Metab 2013;3:3-4. [Crossref] [PubMed]
- Tiong KH, Tan BS, Choo HL, et al. Fibroblast growth factor receptor 4 (FGFR4) and fibroblast growth factor 19 (FGF19) autocrine enhance breast cancer cells survival. Oncotarget 2016;7:57633-50. [PubMed]
- Ahn SM, Jang SJ, Shim JH, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 2014;60:1972-82. [PubMed]
- French DM, Lin BC, Wang M, et al. Targeting FGFR4 inhibits hepatocellular carcinoma in preclinical mouse models. PLoS One 2012;7:e36713. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)


